Abstract
Background
Aluminum (Al) toxicity has limited the productivity and expansion of cereal crops on acid soils; however, a number of plant species or cultivars have developed different strategies for detoxifying aluminum both internally and externally.
Scope
This review focuses on recent progress on molecular mechanisms of Al tolerance in gramineous plants.
Conclusions
A common mechanism in all gramineous plants is the secretion of organic acid anions (citrate and malate) from the roots. Genes belonging to ALMT (for Aluminum-activated malate transporter) and MATE (Multidrug and toxic compound extrusion) family involved in the secretion have been identified in several plant species; however, different plant species show different gene expression patterns including Al-induction, spatial and temporal expression, and tissue localization. Furthermore, the mechanisms regulating the gene expression also differ with plant species, which are achieved by increased tandem repeated element, increase of copy number, insertion of transposon, or alteration of cis-acting element. In addition to these common Al exclusion mechanisms, rice as a highly Al-tolerant species has developed a number of other mechanisms for detoxification of Al. A transcription factor for Al tolerance ART1 identified in rice regulates at least 30 genes implicated in internal and external detoxification of Al. These multiple genes may contribute to the high Al tolerance of rice. In the future, regulation mechanisms of Al-tolerance genes need to be further investigated.
Similar content being viewed by others
Introduction
The family Gramineae (also called Poaceae) includes many important crops such as rice (Oryza sativa), maize (Zea mays), wheat (Triticum aestivum) and barley (Hordeum vulgare). The three most important cereals; rice, wheat and maize directly contribute more than half of all calories consumed by human beings (Awika 2011). Therefore, increasing the production of these cereal crops is a very important issue in terms of food security because the world population is predicted to increase by about two billions by 2050 (FAO 2011) and another one billion tones of cereals will be required to feed the increasing population every year.
Many cereal cultivars with potential high yield have been bred during decades, however, their genetic potential has been limited due to various stresses. For instance, acid soils are widely distributed around the world and these pose multiple abiotic stresses to plants. Acid soils cover 30–40 % of arable land and more than 50 % of potential arable land (von Uexküll and Mutert 1995), where important cereal crops are cultivated. Constraints to production on acid soils are caused by both deficiency of nutrients such as P, Ca and Mg and toxicity of metals such as Mn and Al, but Al toxicity has been considered as the major limiting factor.
Aluminum is the most abundant metal in soil and comprises about 8 % on the elemental basis. Most Al in soil (in the form of aluminum oxides) is insoluble; however, when soil pH drops, Al becomes more soluble. Soluble Al (mainly Al3+) shows phytotoxicity, which rapidly inhibits root elongation at the micromolar level, subsequently affecting the uptake of water and nutrients (Kochian 1995; Ryan et al. 2001; Ma 2007). Aluminum inhibits both cell division and elongation of the roots, with a time- and Al concentration-dependent manner (Kochian 1995; Ma 2000). Exposure to a low Al concentration may only inhibit the cell elongation during a short-period. However, a prolonged treatment with a high Al concentration will inhibit both cell elongation and division. Intensive studies on the mechanism of Al toxicity have been made and many possibilities have been proposed (Horst 1995; Ma 2000; Kochian et al. 2005; Poschenrieder et al. 2008; Horst et al. 2010). It seems that Al targets multiple sites of the roots including cell wall, plasma membrane, mitochondrial and nucleus. Binding of Al to these components affects the function and integrity of the root cells. This is supported by a recent microarray analysis study: a short exposure to Al induced expression (both up and down) of a number of genes (Tsutsui et al. 2012). Interestingly, genes not only in the root tips, the site of Al toxicity, but also those in the mature root zone and above-ground part were also induced although more genes in the root tips were affected (Tsutsui et al. 2012).
On the other hand, there is a wide variation in Al tolerance between species and cultivars within a species. Intensive studies have also been made to understand how these Al-tolerant species or cultivars cope with Al toxicity. Physiological studies have revealed several mechanisms of Al tolerance in different plant species, which include organic acid anions secretion from the roots (Miyasaka et al. 1991; Delhaize et al. 1993; Pellet et al. 1995; Ma et al. 1997b), cell wall modification (Yang et al. 2008; Yang et al. 2011), alkalination of the rhizosphere (Degenhardt et al. 1998) and sequestration of Al into vacuoles (Shen et al. 2002, 2003). A number of loci responsible for Al tolerance have also been identified in different gramineous plants (Wu et al. 2000; Ma et al. 2002; Nguyen et al. 2001, 2003; Ma et al. 2004; Magalhaes et al. 2004; Niedziela et al. 2013). Especially in the last decade, great progresses have been made in identification of genes involved in Al tolerance. In this review, we focus on recent progress on molecular mechanisms of Al tolerance in gramineous plants. For the progress made in other plant species, refer to recent review papers (e.g. Delhaize et al. 2012).
Interspecific and genotypic difference in Al tolerance in gramineous plants
There is a great variation in Al tolerance among gramimeous species and cultivars within a species. In cereal crops, the Al tolerance usually follows rice, rye (Secale cereale) > oat (Avena sativa) > wheat > barley (Bona et al. 1993). However, due to root elongation rates and genotypic differences, it is difficult to directly compare Al tolerance between species.
Genotypic differences in Al tolerance has been reported in many species. In rice, usually japonica cultivars are more tolerant to Al than indica cultivars (Ma et al. 2002). A screening of barley germplasm also showed that there is a large genotypic difference in Al tolerance (Fig. 1; Ma et al. 1997a). Al-tolerant and -sensitive cultivars were also found in wheat, rye, maize and sorghum (Sorghum bicolor) (Rengel and Jurkic 1993; Ryan et al. 1995; Pinto-Carnide and Guedes-Pinto 1999; Pandey et al. 2007; Caniato et al. 2007). These variations have been providing materials for studying Al tolerance mechanisms and identifying Al-tolerance genes as described below.
Physiological mechanisms of Al tolerance in gramineous plants
Physiological studies have revealed that most gramineous species share a common mechanism of Al tolerance; that is secretion of organic acid anions from the roots in response to Al (Ma et al. 2001). However, they differ in the kind of organic acid anions, amount and secretion patterns. Rice (Ma et al. 2002), barley (Zhao et al. 2003), maize (Pellet et al. 1995) and sorghum (Magalhaes et al. 2007) only secrete citrate, but most Al tolerant wheat cultivars secrete malate although some cultivars also secrete citrate (Delhaize et al. 1993; Ryan et al. 2009). Rye (Li et al. 2000), oat (Zheng et al. 1998), triticale (x Triticosecale Wittmark) (Ma et al. 2000) and Yorkshire fog (Holcus lanatus) (Chen et al. 2013) secrete both citrate and malate. Both malate and citrate are able to chelate toxic Al, thereby detoxifying Al in the rhizosphere (Ma et al. 2001).
The amount of Al-induced secretion of organic acid anions also differs with plant species. Usually rye secretes more organic acid anions than wheat and barley (Zhao et al. 2003). Rice only secreted a small amount of citrate although this also contributes to Al tolerance (Ma et al. 2002; Yokosho et al. 2011).
There are two different patterns for organic acid anion secretion (Ma 2000; Ma et al. 2001). In barley and wheat (Pattern I), the secretion occurs immediately after the addition of Al; while in rye, sorghum, rice and maize (Pattern II), there is a delay between the secretion and exposure to Al. These differences are consistent with the expression patterns of Al-tolerance genes involved as described below.
Other physiological mechanisms have also been reported in gramineous plants. For examples, methylation of pectin to reduce Al binding to cell wall in rice (Yang et al. 2008); decreasing the ratio of phospholipids to △5-sterols in the plasma membrane to lower the negative binding site for Al binding in rice (Khan et al. 2009); Al activated exudation of phenolic compounds from maize roots for detoxification of Al in the rhizosphere (Kidd et al. 2001).
Al tolerance genes in gramineous plants
Al tolerance in barley (Ma et al. 2004; Furukawa et al. 2007), wheat (Aniol 1990), sorghum (Magalhaes et al. 2004, 2007) and oat (Nava et al. 2006) is largely controlled by a single dominant gene, while Al tolerance in maize (Pandey et al. 1994; Borrero et al. 1995; Ninamango-Cárdenas et al. 2003; Krill et al. 2010) and rice (Wu et al. 2000; Ma et al. 2002; Nguyen et al. 2001, 2003) is a quantitative trait controlled by multiple genes. Since the first Al-tolerance gene (ALMT1) was identified from wheat (Sasaki et al. 2004), a number of genes involved in Al tolerance have been identified by using different approaches in gramineous plants. Most of them are related to organic acid anion secretion although other Al-tolerance genes also have recently identified in rice.
Genes involved in organic acid anion secretion
ALMT and MATE
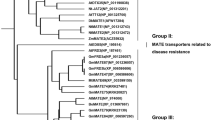
Genes controlling efflux of malate and citrate from the roots have been identified in several gramineous plants. They belong to two different families; ALMT (for Aluminum-activated malate transporter) and MATE/AACT (Multidrug and toxic compound extrusion/Aluminum activated citrate transporter) (Delhaize et al. 2012). TaALMT1 is a plasma membrane-localized protein with six transmembrane domains (Fig. 2c), which mediates the efflux of malate (Sasaki et al. 2004; Yamaguchi et al. 2005). Homologues of TaALMT1 have been identified in rye (ScALMT1, Collins et al. 2008), maize (ZmALMT1 and ZmALMT2, Piñeros et al. 2008; Ligaba et al. 2012), barley (HvALMT1, Gruber et al. 2010) and Yorkshire fog (HlALMT1, Chen et al. 2013) (Fig. 2a), but ZmALMT1, ZmALMT2 and HvALMT1 are not directly involved in the secretion of malate. On the other hand, MATE/AACT is also localized in the plasma membranes usually with nine transmembrane domains (Fig. 2d), but transports citrate (Furukawa et al. 2007). MATE genes have been identified in barley (HvAACT1, Furukawa et al. 2007), sorghum (SbMATE, Magalhaes et al. 2007), rye (ScFRDL2, Yokosho et al. 2010), maize (ZmMATE1, Maron et al. 2010), rice (OsFRDL4, Yokosho et al. 2011) and wheat (TaMATE1B, Tovkach et al. 2013) (Fig. 2b). These genes have been demonstrated to be involved in detoxification of Al toxicity in several species. Knockout or knockdown of these genes results in decreased Al tolerance. On the other hand, over-expression of these genes increases the tolerance to Al toxicity in some cases (Delhaize et al. 2004, 2009; Pereira et al. 2010; Zhou et al. 2013). Interestingly, different species show distinct expression patterns, localization and regulation as detailed below.
Phylogenetic trees and secondary protein structure of ALMT and MATE/AACT in gramineous plants. Peptide sequence alignment was analyzed by Clustal W using default settings (http://www.genome.jp/tools/clustalw/). The phylogenetic tree of ALMT (a) and MATE/AACT (b) was constructed using the Tree View program with the amino acid sequences. Frame marked genes are Al-inducible. The common membrane topology of ALMT (c) and MATE/AACT (d) is predicted by TMHMM server (http://www.cbs.dtu.dk/services/TMHMM/)
Different expression pattern and localization
The expression pattern of ALMT and MATE differs with plant species in terms of induction, spatial and temporal expression. The expression of TaALMT1 (Sasaki et al. 2004), and HvAACT1 (Furukawa et al. 2007), is not induced by Al, whereas that of ScALMT1 (Collins et al. 2008), ScFRDL2 (Yokosho et al. 2010), SbMATE (Magalhaes et al. 2007), OsFRDL4 (Yokosho et al. 2011) and HlALMT1 (Chen et al. 2013) is up-regulated by Al (Fig. 2a, b). Furthermore, the expression of ScALMT1 and OsFRDL4 is up-regulated within hours upon exposure to Al, while that of SbMATE requires few days to be induced (Collins et al. 2008; Yokosho et al. 2011; Magalhaes et al. 2007). These expression patterns are consistent with secretions patterns of organic acid anions as described above. Rapid secretion in Pattern I species does not require gene induction, while gene induction is required for the delayed secretion in Pattern II species.
Most ALMT and MATE/AACT genes are expressed in the root tips. For example, TaALMT1 is highly expressed in the root tips (0–5 mm; Sasaki et al. 2004). High expression of HvAACT1 is also observed in the first 3 mm of barley roots (Fujii et al. 2012). Since root tips are the target site of Al toxicity (Ryan et al. 1993), high expression in this position can efficiently detoxify Al externally. However, some MATE genes are also expressed in the mature root zones. For example, similar expression level of OsFRDL4 was found in both the root tips and mature zones (Yokosho et al. 2011). In an Al-tolerant cultivar of wheat (Carazinho), higher expression of TaMATE1B was found in the basal root region than in the root tips (Tovkach et al. 2013). There are two possibilities for the expression of MATE in the basal root zones. One is that citrate is also secreted from this region to prevent toxic Al into the cells. The other is that these genes may play different role in the mature root zone. This is demonstrated by a recent study showing that HvAACT1 in the mature root region is involved in translocation of Fe from the roots to the shoots (Fujii et al. 2012).
The tissue and cellular localization was only investigated in a limited number of species. In barley, HvAACT1 is localized in the epidermal cells (Furukawa et al. 2007). In rice, OsFRDL4 is expressed in all rice root cells (Yokosho et al. 2011). Recently, SbMATE in sorghum was found to be localized in the epidermal and outer cell cortical cell layers of the DTZ (Sivaguru et al. 2013). The difference in the tissue localization may be associated with the root structures of different plant species.
Regulation mechanism of ALMT and MATE gene expression
The expression level of ALMT and MATE/AACT is higher in Al-tolerant cultivars than in Al-sensitive cultivars, indicating that the Al tolerance is regulated by the expression level. Recently, a few studies have revealed the mechanisms regulating the expression of ALMT and MATE/AACT. They include increased tandem repeated element, increase of copy number, insertion of transposon, and alteration of cis-acting element (Fig. 3, Delhaize et al. 2012). The wheat TaALMT1 promoter is highly polymorphic with eight different alleles described to date. These alleles comprise of single nucleotide polymorphisms (SNPs) and perfect tandem repeats of various sizes. The high expression level of ALMT1 is associated with tandem repeated elements in the promoter region in most Al-tolerant lines (Fig. 3a, Sasaki et al. 2006; Ryan et al. 2010). In sorghum, tourist-like miniature inverted repeat transposable elements (MITEs) occur upstream of the SbMATE gene and the number of these repeats is broadly correlated with the level of SbMATE expression (Fig. 3a, Magalhaes et al. 2007). In rye, the high Al tolerance in Al-tolerant genotype is attributed to the increased genomic copy number of ScALMT1 (Fig. 3b, Collins et al. 2008). Al-tolerant genotypes of rye have five ScALMT1 genes clustered together on chromosome 7R, of which two are highly expressed in the root tip, whereas only two copies in the sensitive genotype, of which only one are highly expressed in the root tip (Collins et al. 2008). Recently, Maron et al. (2013) also found that Al-tolerant cultivars of maize have three copies of ZmMATE1 in the genome, which are identical and part of a tandem triplication (Fig. 3b). This copy number is associated with both gene expression and Al tolerance (Maron et al. 2013). On the other hand, transposon insertion was found to regulate the expression level in barley and some wheat cultivars (Fig. 3c). In barley, a 1-kb insertion (CACTA-like transposon) was found in the 5′ untranslated region (UTR) of the HvAACT1 coding region of Al-tolerant cultivars. This insertion functions as a promoter, which enhances and alters the tissue localization of HvAACT1 (Fujii et al. 2012). Recently, an 11.1-kb transposon-like element (a Sukkula-like transposon) located 25 bp upstream of the start codon was also found to extend TaMATE1B expression to the root apex of Al-tolerant wheat cultivar Carazinho, which secretes citrate constitutively (Tovkach et al. 2013). In an accession of Yorkshire fog well adopted in acid soil, the high expression of HlALMT1 is achieved by increasing the number of cis-acting elements of ART1 in the promoter region (Fig. 3d, Chen et al. 2013). ART1 is a transcription factor for Al tolerance as described below. These findings indicate the expression of ALMT1 and MATE1 is regulated by different ways depending on plant species.
Different regulation mechanisms of Al-tolerance genes involved in organic acid anion secretion in gramineous plants. The high gene expression level is achieved by increase of tandem repeated element (a), insertion of transposon (b), increase of copy number (c), or alteration of cis-acting element (d). Modified according to Delhaize et al. (2012)
Interestingly, in barley, the 1-kb insertion was only detected in some accessions cultivated in acid soil area including Japan, China and Korea, but not in wild barley (Fujii et al. 2012). Three-copy alleles of ZmMATE1 were only found in three cultivars of maize, which share the same geographical origin, in regions of acid soils of the South American tropics (Maron et al. 2013), but not in teosinte (Zea luxurians). The difference in the number of cis-acting elements in Yorkshire fog seems to have occurred over 150 years due to single nucleotide substitutions although this is also a possibility that these SNPs are the result of selection of various alleles on the acidic plots (Chen et al. 2013). These findings suggest that the regulatory mechanism of the gene expression was developed as a result of adaptation to acid soil during relatively short term as proposed by Ryan and Delhaize (2010).
Mechanism of Al-induced activation
Heterologous expression analysis showed that most ALMT and MATE proteins require external Al to activate their function. However, the mechanism underlying Al activation is still poorly understood. Motoda et al. (2007) reported that ALMT1 protein contains six transmembrane domains with the amino and carboxyl termini located on the extracellular side of the plasma membrane. Furuichi et al. (2010) found that three main residues (Glu274, Asp275 and Glu284) in the C-terminal domain of TaALMT1 are involved in the Al-activation response by directly binding Al. However, a recent study showed that the N-domain, which is predicted to form the conductive pathway, can mediate organic anion transport in the absence of the C-domain (Ligaba et al. 2013). Furthermore, it was found that both the N- and C-domains are involved in Al-mediated enhancement of transport activity. It is argued that the C-terminal half is, at least in part, oriented towards the intracellular space, and is thus incapable of direct interaction with extracellular Al (Dreyer et al. 2012; Mumm et al. 2013). In addition, a recent comprehensive phytogenetic analysis showed that these three residues are highly conserved throughout the entire ALMT family, in which some members do not show Al-activation. Information of crystal structure of ALMT1 will be required in order to understand the interaction between Al and the proteins. Although MATE activity was also activated by Al (Furukawa et al. 2007; Yokosho et al. 2011), there is no study on the underlying mechanism.
ART1-mediated high Al tolerance in rice
Among the small grain cereal crops, rice is characterized by superior Al tolerance (Ma et al. 2002). Genetic analysis has shown that Al tolerance in rice is controlled by multiple genes and a number of loci associated with Al tolerance have been detected (Wu et al. 2000; Ma et al. 2002; Nguyen et al. 2001, 2003). By using mutant approach, a transcription factor for Al tolerance, ART1 (Al resistance transcription factor 1) has been identified (Yamaji et al. 2009). ART1 is a C2H2 zinc-finger protein, which is a homolog of STOP1. In Arabidopsis thaliana, STOP1 regulates the expression of AtMATE1 and AtALMT1 (Liu et al. 2009; Sawaki et al. 2009). ART1 binds to the core cis-acting element [GGN(T/g/a/C)V(C/A/g)S(C/G)] that is present in the promoter of ART1 downstream genes (Tsutsui et al. 2011). ART1 regulates at least 31 genes (Yamaji et al. 2009). Functional analysis of some of ART1 downstream genes showed that they are involved in detoxification of Al both externally and internally. Although the expression and localization of ART1 is not induced by Al, the expression of downstream genes is up-regulated by Al within hours (Yamaji et al. 2009). All the proteins encoded are localized at all root cells (Huang et al. 2009, 2012; Xia et al. 2010, 2013; Yokosho et al. 2011; Chen et al. 2012), but play different roles in Al detoxification (Fig. 4). STAR1 and STAR2 encode an ATP-binding domain and a membrane-binding domain, respectively, of a bacterial type ABC transporter (Huang et al. 2009). The STAR1-STAR2 complex localized at the vesicles transports UDP-glucose, which may be involved in cell wall modification, resulting in decreased Al accumulation in the cell wall. Four genes (OsFRDL4, OsCDT3, OsMGT1 and OsNrat1) all encode plasma-membrane-localized proteins, which are involved in Al tolerance. OsFRDL4 described above is responsible for the secretion of citrate in response to Al (Yokosho et al. 2011). Compared with other ART1-regulated genes, the up-regulation of OsFRDL4 by Al is the greatest. OsCDT3, encoding a small cysteine-rich peptide shows binding activity with Al, thereby preventing Al entering into the root cells (Xia et al. 2013). On the other hand, OsMGT1 functions as a Mg transporter (Chen et al. 2012), and up-regulation of OsMGT1 could alleviate internal Al toxicity by enhancing Mg uptake. The involvement of Mg transporter in Al tolerance was also reported in yeast (ALR1 or ALR2) and Arabidopsis (AtMGT1) (MacDiarmid and Gardner 1998; Deng et al. 2006). Nrat1, a member of Nramp family, transports trivalent Al (Xia et al. 2010), which is required for sequestration of Al into the vacuoles for final detoxification. Vacuolar sequestration of Al is mediated by OsALS1, a half-size ABC transporter localized at the tonoplast (Huang et al. 2012). Knockout of any gene described above results in increased Al sensitivity, but at different extent. Among these genes examined, OsFRDL4 showed a good correlation between the expression and Al tolerance (Yokosho et al. 2011). Variation of Nrat1 explained 40 % of Al tolerance in aus subpopulation from Indica varietal group (Famoso et al. 2011). These findings suggest that these two genes may be responsible for genotypic difference in Al tolerance. By contrast, there is no correlation between the expression of STAR1, STAR2, OsMGT1, OsCDT3, OsALS1 and Al tolerance, suggesting that they are involved in basic detoxification of Al in rice. Therefore, the high Al tolerance in rice is achieved by multiple genes involved in both internal and external detoxification.
A homolog of rice ART1 was also identified in Yorkshire fog (Chen et al. 2013). Similar to rice ART1, HlART1 interacts with the promoter region of HlALMT1, suggesting that rice and Yorkshire fog share a similar regulatory pathway. Recently, an ART1 homolog in wheat, TaSTOP1, was cloned (Garcia-Oliveira et al. 2013). The expression of TaSTOP1 is also not induced by Al, but the role of this gene in Al tolerance remains to be investigated further since TaALMT1 is expressed constitutively.
In addition to ART1, recently ASR5 (Abscisic acid, stress and ripening) was reported to be involved in Al tolerance in rice (Arenhart et al. 2013a). ASR5 is localized in both the nucleus and the cytoplasm and acts as a transcription factor, which may regulate the expression of different genes that collectively protect rice cells from Al-induced stress responses. Furthermore, ASR5 protein could bind STAR1 promoter (Arenhart et al. 2013b). However, knockdown of this gene not only resulted in decreased Al tolerance, but also decreased tolerance to other stress such as drought and morphologic changes (Arenhart et al. 2013a). This raises a question whether ASR5 is involved in Al tolerance directly.
Manipulation of Al-tolerance genes
Identification of Al-tolerance genes makes it possible to introduce these genes into Al-sensitive species and cultivars, thereby producing transgenic crops with high Al tolerance. Several attempts have been made, mainly by enhancing organic acid anion secretion. Secretion of organic acid anions from the roots involves synthesis and transport processes. Since organic acids such as malate and citrate are common cellular components with high turnover rates and only a small part of organic acids are secreted in response to Al, the synthesis process is unlikely a limiting factor for secretion of organic acid anions (Ryan et al. 2011). However, several studies have shown that over-expression of genes encoding organic acid synthesis also resulted in increased citrate secretion and Al tolerance (Anoop et al. 2003; de la Fuente et al. 1997; Koyama et al. 2000; Tesfaye et al. 2001), although the same result was not observed in another study (Delhaize et al. 2001). By contrast, over-expression of genes involved in secretion of organic acids has successfully improved the Al tolerance. Over-expression of TaALMT1 in barley, wheat and Arabidopsis resulted in enhanced secretion of malate and Al tolerance (Delhaize et al. 2004; Pereira et al. 2010; Ryan et al. 2011). Introduction of MATE genes also resulted in increased citrate secretion and Al tolerance in tobacco (Nicotiana tabacum) and Arabidopsis (Furukawa et al. 2007; Magalhaes et al. 2007). Recently, it was also found to be effective to increase Al tolerance in barley and wheat by over-expressing HvAACT1 (Zhou et al. 2013). However, it seems that there is a limit to increase Al tolerance by manipulating these genes. For example, transgenic barley carrying HvAACT1 showed lower Al tolerance compared with Al-tolerant barley cultivar, Dayton (Zhou et al. 2013). These findings suggest that there is still potential to enhance Al tolerance by introducing multiple genes rather than a single gene and genes involved in the regulations.
Prospective
Great progresses have been made in understanding molecular mechanism of Al tolerance in gramineous plants during the last decade. It becomes clear that species having multiple Al-tolerance genes like rice is more tolerant to Al than species having single tolerance gene like barley. For the single Al-tolerance gene such as ALMT and MATE/AACT, the expression level determines the Al tolerance. Although the mechanisms regulating the expression level of ALMT and MATE/AACT have been elucidated in some species as described above, the whole picture on how these genes are regulated in different species and cultivars, remains to be further investigated in detail. Recently, Melo et al. (2013) found that introduction of SbMATE into different background resulted in different expression level and Al tolerance, suggesting that SbMATE expression is regulated at multiple levels. It seems that both cis- and trans-acting elements are involved in regulating SbMATE expression.
The transcription factor ART1 plays a central role in Al tolerance in rice and Yorkshire fog (Yamaji et al. 2009; Chen et al. 2013). However, the upstream pathway is still unknown. The expression and localization of ART1 is unaffected by Al (Yamaji et al. 2009), therefore, there must be a process for activation of ART1. It will be interesting to identify ART1-interacting proteins in future. On the other hand, although eight downstream genes of ART1 have been functionally characterized, the role of remaining genes is unknown. Functional characterization of these genes will extend our understanding on high Al tolerance in rice. It will also be interesting to examine whether ART1-mediated Al tolerance is also present in other gramineous species such as barley and wheat, and why some genes such as HvAACT1 and TaALMT1 are not induced by Al. Famoso et al. (2011) detected 46 candidate genes for Al tolerance in rice by GWA (Genome-wide association) analysis. Identification of these QTL genes will provide new insight into molecular mechanisms underlying genotypic difference in Al tolerance.
For breeding Al-tolerant species and cultivars, in addition to transgenic approaches as described above, other approaches such as marker-assisted selection (MAS), genomic selection will also be useful. Introduction of multiple Al-tolerance genes rather than a single gene will be more effective to improve Al tolerance and therefore increase crop production on acid soils.
References
Aniol A (1990) Genetics of tolerance to aluminum in wheat (Triticum aestivum L.). Plant Soil 123:223–227
Anoop VM, Basu U, McCammon MT, McAlister-Henn L, Taylor GJ (2003) Modulation of citrate metabolism alters aluminum tolerance in yeast and transgenic canola overexpressing a mitochondrial citrate synthase. Plant Physiol 132:2205–2217
Arenhart RA, Bai Y, Valter de Oliveira LF et al (2013a) New insights into aluminum tolerance in rice: the ASR5 protein binds the STAR1 promoter and other aluminum-responsive genes. Mol Plant. doi:10.1093/mp/sst160
Arenhart RA, Lima JC, Pedron M et al (2013b) Involvement of ASR genes in aluminum tolerance mechanisms in rice. Plant Cell Environ 36:52–67
Awika JM (2011) Major cereal grains production and use around the world. In advances in cereal science: implications to food processing and health promotion. ACS symposium series; American Chemical Society. Washington DC, pp 1–13
Bona L, Wright RJ, Baligar VC, Matuz J (1993) Screening wheat and other small grains for acid soil tolerance. Landsc Urban Plan 27:175–178
Borrero JC, Pandey S, Ceballos H, Magnavaca R, Bahia AFC (1995) Genetic variances for tolerance to soil acidity in a tropical maize population. Maydica 40:283–288
Caniato FF, Guimarães CT, Schaffert RE, Alves VMC, Kochian LV, Borém A, Klein PE, Magalhaes JV (2007) Genetic diversity for aluminum tolerance in sorghum. Theor Appl Genet 114:863–876
Chen ZC, Yamaji N, Motoyama R, Nagamura Y, Ma JF (2012) Up-regulation of a magnesium transporter gene OsMGT1 is required for conferring aluminum tolerance in rice. Plant Physiol 159:1624–1633
Chen ZC, Yokosho K, Kashino M, Zhao FJ, Yamaji N, Ma JF (2013) Adaptation to acidic soil is achieved by increased cis-acting element numbers regulating ALMT1 expression in Holcus lanatus. Plant J 76:10–23
Collins NC, Shirley NJ, Saeed M, Pallotta M, Gustafson JP (2008) An ALMT1 gene cluster controlling Aluminum tolerance at the Alt4 locus of rye (Secale cereale L.). Genetics 179:669–682
De la Fuente JM, Ramírez-Rodríguez V, Cabrera-Ponce JL, Herrera-Estrella L (1997) Aluminum tolerance in transgenic plants by alteration of citrate synthesis. Science 276:1566–1568
Degenhardt J, Larsen PB, Howell SH, Kochian LV (1998) Aluminum resistance in the Arabidopsis mutant alr-104 is caused by an aluminum-induced increase in rhizosphere pH. Plant Physiol 117:19–27
Delhaize E, Ryan PR, Randall PJ (1993) Aluminum tolerance in wheat (Triticum aestivum L.). II. Aluminum-stimulated excretion of malic acid from root apices. Plant Physiol 103:695–702
Delhaize E, Hebb DM, Ryan PR (2001) Expression of a Pseudomonas aeruginosa citrate synthase gene in tobacco is not associated with either ehnhanced citrate accumulation or efflux. Plant Physiol 125:2059–2067
Delhaize E, Ryan PR, Hebb DM, Yamamoto Y, Sasaki T, Matsumoto H (2004) Engineering high-level aluminum tolerance in barley with the ALMT1 gene. Proc Natl Acad Sci U S A 101:15249–15254
Delhaize E, Taylor P, Hocking PJ, Simpson RJ, Ryan PR, Richardson AE (2009) Transgenic barley (Hordeum vulgare L.) expressing the wheat aluminum resistance gene (TaALMT1) shows enhanced phosphorus nutrition and grain production when grown on an acid soil. Plant Biotechnol J 7:391–400
Delhaize E, Ma JF, Ryan PR (2012) Transcriptional regulation of aluminium tolerance genes. Trends Plant Sci 17:341–348
Deng W, Luo K, Li D, Zheng X, Wei X, Smith W, Thammina C, Lu L, Li Y, Pei Y (2006) Overexpression of an Arabidopsis magnesium transport gene, AtMGT1, in Nicotiana benthamiana confers Al tolerance. J Exp Bot 57:4235–4243
Dreyer I, Gomez-Porras JL, Riaño-Pachón DM, Hedrich R, Geiger D (2012) Molecular evolution of slow and quick anion channels. Front Plant Sci 3:263
Famoso AN, Zhao K, Clark RT, Tung CW, Wright MH, Bustamante C, Kochian LV, McCouch SR (2011) Genetic architecture of aluminum tolerance in rice (Oryza Sativa) determined through genome-wide association analysis and QTL mapping. PLoS Genet 7:e1002221
Food and Agriculture Organization (FAO) (2011) World Livestock 2011. Livestock in food security. Rome
Fujii M, Yokosho K, Yamaji N, Saisho D, Yamane M, Takahashi H, Sato K, Nakazono M, Ma JF (2012) Acquisition of aluminium tolerance by modification of a single gene in barley. Nat Commun 3:713. doi:10.1038/ncomms1726
Furuichi T, Sasaki T, Tsuchiya Y, Ryan PR, Delhaize E, Yamamoto Y (2010) An extracellular hydrophilic carboxy-terminal domain regulates the activity of TaALMT1, the aluminum-activated malate transport protein of wheat. Plant J 64:47–55
Furukawa J, Yamaji N, Wang H, Mitani N, Murata Y, Sato K, Katsuhara M, Takeda K, Ma JF (2007) An aluminum-activated citrate transporter in barley. Plant Cell Physiol 48:1081–1091
Garcia-Oliveira AL, Benito C, Prieto P, de Andrade MR, Rodriques-Pousada C, Guedes-Pinto H, Martins-Lopes P (2013) Molecular characterization of TaSTOP1 homoeologues and their response to aluminium and proton (H+) toxicity in bread wheat (Triticum aestivum L.). BMC Plant Biol 13:134
Gruber BD, Ryan PR, Richardson AE, Tyerman SD, Ramesh S, Hebb DM, Howitt SM, Delhaize E (2010) HvALMT1 from barley is involved in the transport of organic anions. J Exp Bot 61:1455–1467
Horst WJ (1995) The role of the apoplast in aluminum toxicity and resistance of higher plants: a review. Z Pflanzenernähr Bodenkd 158:419–428
Horst WJ, Wang Y, Eticha D (2010) The role of the root apoplast in aluminium-induced inhibition of root elongation and in aluminium resistance of plants: a review. Ann Bot 185–197
Huang CF, Yamaji N, Mitani N, Yano M, Nagamura Y, Ma JF (2009) A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 21:655–667
Huang CF, Yamaji N, Chen Z, Ma JF (2012) A tonoplast-localized half-size ABC transporter is required for internal detoxification of aluminum in rice. Plant J 69:857–867
Khan MS, Tawaraya K, Sekimoto H et al (2009) Relative abundance of delta5-sterols in plasma membrane lipids of root-tip cells correlates with aluminum tolerance of rice. Physiol Plant 135:73–83
Kidd PS, Llugany M, Poschenrieder C, Gunsé B, Barceló J (2001) The role of root exudates in aluminum resistance and silicon-induced amelioration of aluminum toxicity in three varieties of maize (Zea mays L.). J Exp Bot 52:1339–1352
Kochian LV (1995) Cellular mechanisms of aluminum toxicity and resistance in plants. Annu Rev Plant Physiol Plant Mol Biol 46:237–260
Kochian LV, Piñeros MA, Hoekenga OA (2005) The physiology, genetics and molecular biology of plant aluminum resistance and toxicity. Plant Soil 274:175–195
Koyama H, Kawamura A, Kihara T, Hara T, Takita E, Shibata D (2000) Overexpression of mitochondrial citrate synthase in Arabidopsis thaliana improved growth on a phosphorus-limited soil. Plant Cell Physiol 41:1030–1037
Krill AM, Kirst M, Kochian LV, Buckler ES, Hoekenga OA (2010) Association and linkage analysis of aluminum tolerance genes in maize. PLoS ONE 5:e9958
Li XF, Ma JF, Matsumoto H (2000) Pattern of aluminum-induced secretion of organic acids differs between rye and wheat. Plant Physiol 123:1537–1543
Ligaba A, Maron L, Shaff J, Kochian L, Piñeros M (2012) Maize ZmALMT2 is a root anion transporter that mediates constitutive root malate efflux. Plant Cell Environ 35:1185–1200
Ligaba A, Dreyer I, Margaryan A, Schneider DJ, Kochian L, Piñeros M (2013) Functional, structural and phylogenetic analysis of domains underlying the Al sensitivity of the aluminum-activated malate/anion transporter, TaALMT1. Plant J 76:766–780
Liu J, Magalhaes JV, Shaff J, Kochian LV (2009) Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J 57:389–399
Ma JF (2000) Role of organic acids in detoxification of aluminum in higher plants. Plant Cell Physiol 41:384–390
Ma JF (2007) Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int Rev Cytol 264:225–252
Ma JF, Zheng SJ, Li XF, Takeda K, Matsumoto H (1997a) A rapid hydroponic screening for aluminium tolerance in barley. Plant Soil 191:133–137
Ma JF, Zheng SJ, Hiradate S, Matsumoto H (1997b) Detoxifying aluminium with buckwheat. Nature 390:569–570
Ma JF, Taketa S, Yang ZM (2000) Aluminum tolerance genes on the short arm of chromosome 3R are linked to organic acid release in triticale. Plant Physiol 122:687–694
Ma JF, Ryan PR, Delhaize E (2001) Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci 6:273–278
Ma JF, Shen R, Zhao Z, Wissuwa M, Takeuchi Y, Ebitani T, Yano M (2002) Response of rice to Al stress and identification of quantitative trait loci for Al tolerance. Plant Cell Physiol 43:652–659
Ma JF, Nagao S, Sato K, Ito H, Furukawa J, Takeda K (2004) Molecular mapping of a gene responsible for Al-activated secretion of citrate inbarley. J Exp Bot 55:1335–1341
MacDiarmid CW, Gardner RC (1998) Overexpression of the Saccharomyces cerevisiae magnesium transport system confers resistance to aluminum ion. J Biol Chem 273:1727–1732
Magalhaes JV, Garvin DF, Wang Y, Sorrells ME, Klein PE, Schaffert RE, Li L, Kochian LV (2004) Comparative mapping of a major aluminum tolerance gene in sorghum and other species in the Poaceae. Genetics 167:1905–1914
Magalhaes JV, Liu J, Guimarães CT et al (2007) A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat Genet 39:1156–1161
Maron LG, Piñeros MA, Guimarães CT, Magalhaes JV, Pleiman JK, Mao C, Shaff J, Belicuas SNJ, Kochian LV (2010) Two functionally distinct members of the MATE (multi-drug and toxic compound extrusion) family of transporters potentially underlie two major aluminum tolerance QTLs in maize. Plant J 61:728–740
Maron LG, Guimarães CT, Kirst M et al (2013) Aluminum tolerance in maize is associated with higher MATE1 gene copy number. Proc Natl Acad Sci U S A 110:5241–5246
Melo JO, Lana UG, Piñeros MA et al (2013) Incomplete transfer of accessory loci influencing SbMATE expression underlies genetic background effects for aluminum tolerance in sorghum. Plant J 73:276–288
Miyasaka SC, Buta JG, Howell RK, Foy CD (1991) Mechanism of aluminum tolerance in snapbeans. Root exudation of citric acid. Plant Physiol 96:737–743
Motoda H, Sasaki T, Kano Y, Ryan PR, Delhaize E, Matsumoto H, Yamamoto Y (2007) The membrane topology of ALMT1, an aluminum-activated malate transport protein in wheat (Triticum aestivum). Plant Signal Behav 2:467–472
Mumm P, Imes D, Martinoia E, Al-Rasheid KA, Geiger D, Marten I, Hedrich R (2013) C-terminus-mediated voltage gating of Arabidopsis guard cell anion channel QUAC1. Mol Plant 6:1550–1563
Nava LC, Delatorre CA, de Lima Duarte IT, Pacheco MT, Federizzi LC (2006) Inheritance of aluminum tolerance and its effects on grain yield and grain quality in oats (Avena sativa L.). Euphytica 148:353–358
Nguyen VT, Burow MD, Nguyen HT, Le BT, Le TD, Paterson AH (2001) Molecular mapping of genes conferring aluminum tolerance in rice (Oryza sativa L.). Theor Appl Genet 102:1002–1010
Nguyen BD, Brar DS, Bui BC, Nguyen TV, Pham LN, Nguyen HT (2003) Identification and mapping of the QTL for aluminum tolerance introgressed from the new source, Oryza rufipogon Griff., into indica rice (Oryza sativa L.). Theor Appl Genet 106:583–593
Niedziela A, Bednarek PT, Labudda M, Mańkowski DR, Aniol A (2013) Genetic mapping of a 7R Al tolerance QTL in triticale (x Triticosecale Wittmack). J Appl Genet. doi:10.1007/s13353-013-0170-0
Ninamango-Cárdenas FE, Guimaraes CT, Martins PR, Parentoni SN, Carneiro NP, Lopes MA, Moro JR, Paiva E (2003) Mapping QTLs for aluminum tolerance in maize. Euphytica 130:223–232
Pandey S, Ceballos H, Magnavaca R et al (1994) Genetics of tolerance to soil acidity in tropical maize. Crop Sci 34:1511–1514
Pandey S, Alberto L, Leon N et al (2007) Breeding maize for tolerance to soil acidity. Plant Breed Rev 28:59–100
Pellet DM, Grunes DL, Kochian LV (1995) Organic acid exudation as an aluminum-tolerance mechanism in maize (Zea mays L.). Planta 196:788–795
Pereira JF, Zhou G, Delhaize E, Richardson T, Zhou M, Ryan PR (2010) Engineering greater aluminium resistance in wheat by over-expressing TaALMT1. Ann Bot 106:205–214
Piñeros MA, Cançado GMA, Maron LG, Lyi SM, Menossi M, Kochian LV (2008) Not all ALMT1-type transporters mediate aluminum-activated organic acid responses: the case of ZmALMT1 – an anion-selective transporter. Plant J 53: 352–367
Pinto-Carnide O, Guedes-Pinto H (1999) Aluminium tolerance variability in rye and wheat Portuguese germplasm. Genet Resour Crop Evol 46:81–85
Poschenrieder C, Gunsé B, Corrales I, Barceló J (2008) A glance into aluminum toxicity and resistance in plants. Sci Total Environ 400:356–368
Rengel Z, Jurkic V (1993) Evaluation of Triticum aestivum germplasm from Croatia and Yugoslavia for aluminum tolerance. Euphytica 66:111–116
Ryan PR, Delhaize E (2010) The convergent evolution of aluminum resistance in plants exploits a convenient currency. Funct Plant Biol 37:275–284
Ryan PR, Ditomaso JM, Kochian LV (1993) Aluminium toxicity in roots: an investigation of spatial sensitivity and the role of the root cap. J Exp Bot 44:437–446
Ryan PR, Delhaize E, Randall PJ (1995) Malate efflux from root apices and tolerance to aluminum are highly correlated in wheat. Aust J Plant Physiol 22:531–536
Ryan PR, Delhaize E, Jones DL (2001) Function and mechanism of organic anion exudation from plant roots. Annu Rev Plant Physiol Plant Mol Biol 52:527–560
Ryan PR, Raman H, Gupta S, Horst WJ, Delhaize E (2009) A second mechanism for aluminum resistance in wheat relies on the constitutiveefflux of citrate from roots. Plant Physiol 149:340–351
Ryan PR, Raman H, Gupta S, Sasaki T, Yamamoto Y, Delhaize E (2010) The multiple origins of aluminum resistance in hexaploid wheat include Aegilops tauschii and more recent cis mutations to TaALMT1. Plant J 64:446–455
Ryan PR, Tyerman SD, Sasaki T, Furuichi T, Yamamoto Y, Zhang WH, Delhaize E (2011) The identification of aluminium-resistance genes provides opportunities for enhancing crop production on acid soils. J Exp Bot 62:9–20
Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ, Ryan PR, Delhaize E, Matsumoto H (2004) A wheat gene encoding an aluminum-activated malate transporter. Plant J 37:645–653
Sasaki T, Ryan PR, Delhaize E, Hebb DM, Ogihara Y, Kawaura K, Noda K, Kojima T, Toyoda A, Matsumoto H, Yamamoto Y (2006) Sequence upstream of the wheat (Triticum aestivum L.) ALMT1 gene and its relationship to aluminum resistance. Plant Cell Physiol 47:1343–1354
Sawaki Y, Iuchi S, Kobayashi Y, Kobayashi Y, Ikka T, Sakurai N, Fujita M, Shinozaki K, Shibata D, Kobayashi M, Koyama H (2009) STOP1 regulates multiple genes that protect Arabidopsis from proton and aluminum toxicities. Plant Physiol 150:281–294
Shen R, Ma JF, Kyo M, Iwashita T (2002) Compartmentation of aluminium in leaves of an Al-accumulator, Fagopyrum esculentum Moench. Planta 215:394–398
Shen R, Iwashita T, Ma JF (2003) Form of Al changes with A1 concentration in leaves of buckwheat. J Exp Bot 55:131–136
Sivaguru M, Liu J, Kochian LV (2013) Targeted expression of SbMATE in the root distal transition zone is responsible for sorghum aluminum resistance. Plant J 76:297–307
Tesfaye M, Temple SJ, Allan DL, Vance CP, Samac DA (2001) Overexpression of malate dehydrogenase in transgenic alfalfa enhances organic acid synthesis and confers tolerance to aluminum. Plant Physiol 127:1836–1844
Tovkach A, Ryan PR, Richardson AE, Lewis DC, Rathjen TM, Ramesh S, Tyerman SD, Delhaize E (2013) Transposon-mediated alteration of TaMATE1B expression in wheat confers constitutive citrate efflux from root apices. Plant Physiol 161:880–892
Tsutsui T, Yamaji N, Ma JF (2011) Identification of a cis-acting element of ART1, a C2H2-type zinc-finger transcription factor for aluminum tolerance in rice. Plant Physiol 156:925–931
Tsutsui T, Yamaji N, Huang CF, Motoyama R, Nagamura Y, Ma JF (2012) Comparative genome-wide transcriptional analysis of Al-responsive genes reveals novel Al tolerance mechanisms in rice. PLoS ONE 7:e48197
Von Uexküll HR, Mutert E (1995) Global extent, development and economic impact of acid soils. Plant Soil 171:1–15
Wu P, Liao CY, Hu B, Yi KK, Jin WZ, Ni JJ, He C (2000) QTLs and epistasis for aluminum tolerance in rice (Oryza sativa L.) at different seedling stages. Theor Appl Genet 100:1295–1303
Xia JX, Yamaji N, Kasai T, Ma JF (2010) Plasma membrane localized transporter for aluminum in rice. Proc Natl Acad Sci U S A 107:18381–18385
Xia JX, Yamaji N, Ma JF (2013) A plasma membrane-localized small peptide is involved in rice aluminum tolerance. Plant J 76:345–355
Yamaguchi M, Sasaki T, Sivaguru M, Yamamoto Y, Osawa H, Ahn SJ, Matsumoto H (2005) Evidence for the plasma membrane localization of Al-activated malate transporter (ALMT1). Plant Cell Physiol 46:812–816
Yamaji N, Huang CF, Nagao S, Yano M, Sato Y, Nagamura Y, Ma JF (2009) A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 21:3339–3349
Yang JL, Li YY, Zhang YJ, Zhang SS, Wu YR, Wu P, Zheng SJ (2008) Cell wall polysaccharides are specifically involved in the exclusion of aluminum from the rice root apex. Plant Physiol 146:602–611
Yang JL, Zhu XF, Peng YX, Zheng C, Li GX, Liu Y, Shi YZ, Zheng SJ (2011) Cell wall hemicelluloses contributes significantly to aluminum adsorption and root growth in Arabidopsis. Plant Physiol 155:1885–1892
Yokosho K, Yamaji N, Ma JF (2010) Isolation and characterization of two MATE genes in rye. Funct Plant Biol 37:296–303
Yokosho K, Yamaji N, Ma JF (2011) An Al-inducible MATE gene is involved in external detoxification of Al in rice. Plant J 68:1061–1069
Zhao Z, Ma JF, Sato K, Takeda K (2003) Differential Al resistance and citrate secretion in barley (Hordeum vulgare L.). Planta 217:794–800
Zheng S, Ma JF, Matsumoto H (1998) Continuous secretion of organic acids is related to aluminum resistance during relatively long-term exposure to aluminum stress. Physiol Plant 103:209–214
Zhou GF, Delhaize E, Zhou M, Ryan PR (2013) The barley MATE gene, HvAACT1, increases citrate efflux and Al tolerance when expressed in wheat and barley. Ann Bot 112:603–612
Acknowledgments
Part researches cited in this paper were supported by a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan (No. 22119002 to J. F. M.), Ohara foundation for agriculture research, by the Nation Key Basic Research Program of China (No.2014CB441000 to R. F. S) and the Natural Science Foundation of China (No.41025005).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Philippe Hinsinger.
Rights and permissions
About this article
Cite this article
Ma, J.F., Chen, Z.C. & Shen, R.F. Molecular mechanisms of Al tolerance in gramineous plants. Plant Soil 381, 1–12 (2014). https://doi.org/10.1007/s11104-014-2073-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-014-2073-1