Abstract
Background and aims
Sorghum is the second most cultivated crop in Africa and is a staple food source in many African communities. Exploiting the associated plant growth-promoting bacteria (PGPB) has potential as an agricultural biotechnology strategy to enhance sorghum growth, yield and nutritional properties. Therefore this study aimed to evaluate factors that shape bacterial communities associated with sorghum farmed in South Africa, and to detect bacteria consistently associated with sorghum which may impart PGP activities.
Methods
Terminal-Restriction Fragment Length Polymorphism (T-RFLP) was used to assess factors that potentially shape rhizospheric (rhizosphere and rhizoplane) and endophytic (root, shoot, stem) bacterial communities associated with South African sorghum, and together with Denaturing Gradient Gel Electrophoresis (DGGE) to identify consistently sorghum-associated bacterial taxa.
Results
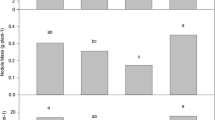
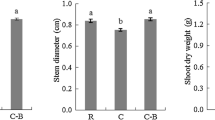
The sorghum rhizospheric communities were less variable than the endophytic ones. Geographical location was the main driver in describing bacterial community assemblages found in rhizospheric sorghum-linked niches, with total NO3-N, NH4-N, nitrogen, carbon, pH and, to a lesser extent, clay content identified as the main abiotic factors shaping sorghum-associated soil communities. Endophytic communities presented rather stochastic assemblages, with pH being the main variable explaining their structures. Despite community variations, specific bacterial taxa were consistently detected in sorghum-created rhizospheric and endophytic environments, irrespective of environmental factor effects.
Conclusions
Soil structure and composition, which are influenced by agricultural practices, played major roles in shaping sorghum-associated edaphic bacterial communities. In contrast, endophytic bacterial communities displayed more variation. Nevertheless, potentially agronomically relevant (cyano)bacterial taxa constantly associated with sorghum were identified which is suggestive of their deterministic recruitment.





Similar content being viewed by others
References
Adams DG, Duggan PS (2012) Signaling in cyanobacteria–plant symbioses. In: Perotto S, Baluška F (eds) Signaling and communication in plant symbiosis. Springer, Berlin Heidelberg, pp 93–121
Bai Y, Aoust FD, Smith DL, Driscoll BT (2002) Isolation of plant growth promoting Bacillus strains from soybean root nodules. Can J Microbiol 48:230–238
Berg G (2009) Plant–microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl Microbiol Biotechnol 84:11–18
Blackwood CB, Hudleston D, Zak DR, Buyer JS (2007) Interpreting ecological diversity indices applied to terminal restriction fragment length polymorphism data: insights from simulated microbial communities. Appl Environ Microbiol 73:5276–5283
Budi SW, Van Tuinen D, Martinotti G, Gianinazzi S (1999) Isolation from the Sorghum bicolor mycorrhizosphere of a bacterium compatible with arbuscular mycorrhiza development and antagonistic towards soilborne fungal pathogens. Appl Environ Microbiol 65:5148–5150
Carson JK, Rooney D, Gleeson DB, Clipson N (2007) Altering the mineral composition of soil causes a shift in microbial community structure. FEMS Microbiol Ecol 61:414–423
Clarke K (1993) Non-parametric multivariate analysis of changes in community structure. Aust J Ecol 18:117–143
Clarke KR, Gorley RN (2006) PRIMER v6: user manual. Plymouth Marine Laboratory, Plymouth
Cocking EC (2003) Endophytic colonization of plant roots by nitrogen-fixing bacteria. Plant Soil 252:169–175
Coelho MRR, de Vos M, Carneiro NP, Marriel IE, Paiva E, Seldi L (2008) Diversity of nifH gene pools in the rhizosphere of two cultivars of sorghum (Sorghum bicolor) treated with contrasting levels of nitrogen fertilizer. FEMS Microbiol Lett 279:15–22
Coelho MRR, Carneiro NP, Marriel IE, Seldin L (2009) Molecular detection of nifH gene-containing Paenibacillus in the rhizosphere of sorghum (Sorghum bicolor) sown in Cerrado soil. Lett Appl Microbiol 48:611–617
Compant S, Duffy B, Nowak J, Clemet C, Barka EA (2005) Use of plant-growth promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl Environ Microbiol 71:4951–4959
Culman SW, Bukowski R, Gauch HG, Cadillo-Quiroz H, Buckley DH (2009) T-REX: software for the processing and analysis of T-RFLP data. BMC Bioinforma 10:171
Dicko MH, Gruppen H, Traore AS, Voragen AGJ, van Berkel WJH (2006) Sorghum grain as human food in Africa: relevance of content of starch and amylase activities. Afr J Biotechnol 5:384–395
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fierer N, Jackson RB (2005) The diversity and biogeography of soil bacterial communities. PNAS 103:626–631
Food and Agricultural Organization (FAO) (1995) FAO food and nutrition series, no. 27 Sorghum and millet in human nutrition. http://www.fao.org/docrep/T0818E/T0818E00.htm
Food and Agricultural Organization/World Food Production (FAO/WFP) (2010) Crop and food security assessment mission to Mozambique. http://www.fao.org/docrep/012/ak350e/ak350e00.htm
Franche C, Lindström K, Elmerich C (2009) Nitrogen-fixing bacteria associated with leguminous and non-leguminous plants. Plant Soil 321:35–59
Funnel-Harris DL, Sattler SE, Pedersen JF (2013) Characterization of fluorescent Pseudomonas spp. Associated with roots and soil of two sorghum genotypes. Eur J Plant Pathol. doi:10.1007/s10658-013-0179-6
Girvan MS, Bullimore J, Pretty JN, Osborn MA, Ball AS (2003) Soil type is the primary determinant of the composition of the total and active bacterial communities in arable soils. Appl Environ Microbiol 69:1800–1809
Gottel NR, Castro HF, Kerley M et al (2011) Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types. Appl Environ Microbiol 77:5934–5944
Hafeez FY, Yasmin S, Ariani D, Mehboob-ur-Rahman Zafar Y, Malik KA (2006) Plant growth-promoting bacteria as biofertilizer. Agron Sustain Dev 26:143–150
Hardoim PR, van Overbeek LS, van Elsas JD (2008) Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol 16:463–471
Hartmann A, Schimd M, van Tuinen D, Berg G (2009) Plant-driven selection of microbes. Plant Soil 321:235–257
Hinsinger P, Bengough AG, Vetterlein D, Young IM (2009) Rhizosphere: biophysics, biogeochemistry and ecological relevance. Plant Soil 321:117–152
Hirel B, Le Gouis J, Ney B, Gallais A (2007) The challenge of improving nitrogen use efficiency in crop plants: towards a more central role for genetic variability and quantitative genetics within integrated approaches. J Exp Bot 58:2369–2387
Kevin Vessey J (2003) Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 255:571–586
Lauber C, Knight R, Hamady M, Fierer N (2009) Soil pH as a predictor of soil bacterial community structure at the continental scale: a pyrosequencing-based assessment. Appl Environ Microbiol 75:5111–5120
Loiret F, Ortega E, Kleiner D, Ortega-Rodes P, Rodes R, Dong Z (2004) A putative new endophytic nitrogen-fixing bacterium Pantoea sp. from sugarcane. J Appl Microbiol 97:504–511
Lugtenberg B, De Weger L, Bennett J (1991) Microbial stimulation of plant growth and protection from disease. Curr Opin Biotechnol 2:457–464
Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, Wade WG (1998) Design and evaluation of useful bacterial primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol 64:795–799
Martiny JBH, Bohannan BJM, Brown JH et al (2006) Microbial biogeography: putting microorganisms on the map. Nat Rev 4:102–112
Mendes R, Pizzirani-Kleiner A, Araujo W, Raaijmarkers J (2007) Diversity of cultivated endophytic bacteria from sugarcane: genetic and biochemical characterization of Burkholderia cepacia complex isolates. Appl Environ Microbiol 73:7259–7267
Morgan JAW, White PJ (2005) Biological costs and benefits to plant-microbe interactions in the rhizosphere. J Exp Bot 56:1729–1739
Murray M, Thompson W (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, New York
Nijland R, Burgess JG, Errington J, Veening J-W (2010) Transformation of environmental Bacillus subtilis isolates by transiently inducing genetic competence. PLoS One 5:e9724. doi:10.1371/journal.pone.0009724
Nocker A, Burr M, Camper AK (2007) Genotypic microbial community profiling: a critical technical review. Microb Ecol 54:276–289
Non-Affiliated Soil Analysis Work Committee (1990) Handbook of standard soil testing methods for advisory purposes, vol 160. Soil Science Society of South Africa, Pretoria
Panchal H, Ingle S (2011) Isolation and characterization of endophytes from the root of the medicinal plant Chlorophytum borivilianum (Safed musli). J Adv Dev Res 2:205–209
Pedersen WL, Chakrabarty K, Klucas RV, Vidave AK (1978) Nitrogen fixation (acetylene reduction) associated with roots of winter wheat and sorghum in Nebraska. Appl Environ Microbiol 35:129–135
Reysenbach A-L, Pace NR (1995) In: Robb FT, Place AR (eds) Archaea: a laboratory. Manual – thermophiles. Cold Spring Harbour Laboratory Press, New York, pp 101–107
Rodriguez-Caballero A, Ramond J-B, Welz PJ, Cowan DA, Odlare M, Burton SG (2012) Treatment of high ethanol concentration wastewater by biological sand filters: enhanced COD removal and bacterial community dynamics. J Environ Manag 109:54–60
Roesch LFW, Camargo FAO, Bento FM, Triplett EW (2008) Biodiversity of diazotrophic bacteria within the soil, root and stem of field-grown maize. Plant Soil 302:91–104
Rosenblueth M, Martinez-Romero E (2006) A review. Bacterial endophytes and their interactions with hosts. Mol Plant Microbe Interact 19:827–837
Rout ME, Chrzanowski TH (2009) The invasive Sorghum halepense harbors endophytic N2-fixing bacteria and alters soil biogeochemistry. Plant Soil 315:163–172
Saharan BS, Nehra V (2011) Plant growth promoting rhizobacteria: a critical review. Life Sci Med Res 21:1–30
Saito A, Ikeda S, Ezura H, Minamisawa K (2007) A minireview. Microbial community analysis of the phytosphere using culture-independent methodologies. Microbes Environ 22:93–105
Schenk PM, Carvalhais LC, Kazan K (2012) Unraveling plant–microbe interactions: Can multi-species transcriptomics help? Trends Biotechnol 30:177–184
Seghers D, Wittebolle L, Top EM, Verstraete W, Siciliano SD (2004) Impact of agricultural practices on the Zea mays L. endophytic community. Appl Environ Microbiol 70:1475–1482
Sercu B, van de Werfhorst LC, Murray JLS, Holden PA (2011) Cultivation-independent analysis of bacteria in IDEXX quanti-tray/2000 fecal indicator assays. Appl Environ Microbiol 77:627–633
Shyu C, Soule T, Bent SJ, Foster JA, Forney LJ (2007) MiCA: a web-based tool for the analysis of microbial communities based on terminal-restriction fragment length polymorphisms of 16S and 18S rRNA genes. J Microb Ecol 53:562–570
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Taylor JRN (2003) Overview: importance of sorghum in Africa. http://www.afripro.org.uk/papers/Paper01Taylor.pdf
Terakado-Tonooka J, Ohwaki Y, Yamakawa H, Tanaka F, Yoneyama T, Fujihara S (2008) Expressed nifH genes of endophytic bacteria detected in field-grown sweet patatoes (Ipoema batatas L.). Microbes Environ 23:89–93
Tyson GW, Baker BJ, Alle EE, Hugenholtz P, Banfield JF (2005) Genome-directed isolation of the key nitrogen fixer Leptospirillum ferrodiazotrophum sp. nov. from an acidic microbial community. Appl Environ Microbiol 71:6319–6324
van Felten A, Meyer JB, Défago G, Maurhofer M (2011) Novel T-RFLP method to investigate six main groups of 2,4-diacetylphloroglucinol-producing pseudomonads in environmental samples. J Microbiol Methods 84:379–387
Zinniel DK, Lambrecht P, Beth Harris N, Feng Z, Kuczmarski D, Higley P, Ishimaru CA, Arunakumari A, Barletta RG, Vidaver AK (2002) Isolation and characterization of endophytic colonizing bacteria from agronomic crops and prairie plants. Appl Environ Microbiol 68:2198–2208
Acknowledgments
We thank the South African National Research Foundation (NRF) for funding the study. We would also like to acknowledge Dr Shagi from the North West academic farm of the ARC (Agricultural Research Council), Mr Osterhaizen (Free State farmer) and the community farm at Jane Furst (Limpopo) for allowing us to sample sorghum. We thank anonymous reviewers who suggested modifications that greatly improved the manuscript. J-BR holds a NRF Free-standing Postdoctoral Fellowship from the National Research Foundation of South Africa.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Paul Bodelier.
Rights and permissions
About this article
Cite this article
Ramond, JB., Tshabuse, F., Bopda, C.W. et al. Evidence of variability in the structure and recruitment of rhizospheric and endophytic bacterial communities associated with arable sweet sorghum (Sorghum bicolor (L) Moench). Plant Soil 372, 265–278 (2013). https://doi.org/10.1007/s11104-013-1737-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-013-1737-6