Abstract
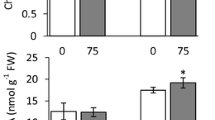
The effects of ionic stress on the physiology and gene expression of two rice genotypes (IR4630 and IR15324) that differ in salt tolerance, were investigated by evaluating changes in the biomass, Na+ and K+ concentrations and applying the cDNA-AFLP technique to highlight changes in gene expression. Over 8 days of salinisation, the effect of NaCl on the reduction of biomass (dry weight) was apparent from 24 h after salinisation (the first time point), indicating that the consequences of the build up of Na+ (and Cl-) in the leaves of both lines was rapid. Furthermore, root growth of IR15324 was much more sensitive to salt than that of IR4630 (the reduction in root dry weight compared to non-salinised plants was three times greater in IR15324 than IR4630). The two rice lines also differed in their Na+ accumulation in saline conditions, a difference that was more marked in the shoots, particularly at the final harvest, than in the roots. Under salt stress, the K+ content (µmol/shoot) increased over four successive harvests (24, 48, 96, 192 h) in both lines, but was always greater in IR4630 than in IR15324: differences in Na+/K+ ratio appear to be an important determinant of salt tolerance in rice. To separate osmotic from ionic effects of salt, mannitol was applied as a non-ionic osmoticum at an osmotic potential estimated to be equivalent to 50 mM NaCl. Messenger RNA was sampled at 0.5, 6, 24, 48 and 192 hours after salinisation. Several products (AFLP-bands) were detected, which were upregulated in the response to ionic effects of salt in the tolerant line (IR4630) and not expressed in the sensitive line (IR15324). Bioinformatic analysis indicated three of these AFLP-bands have a high-degree of sequence similarity with the genes encoding a proline rich protein, senescence associated protein and heat-shock protein. The data are novel in that they differentially highlight changes induced by the ionic rather than osmotic effects of salt and in a tolerant rather than a sensitive genotype. The possible roles of the products of these genes are discussed.






Similar content being viewed by others
Abbreviations
- AFLP:
-
Amplified fragment length polymorphism
- ATP:
-
adenosine tri-phosphate
- cDNA:
-
complementary DNA
- dNTP:
-
deoxynucleotide tri-phosphate
- ds-cDNA:
-
double stranded cDNA
- HSP:
-
heat shock protein
- PAR:
-
photosynthetically active radiation
- PEG:
-
polyethylene glycol
- PRP:
-
proline-rich protein
- RT-PCR:
-
reverse-transcription polymerase chain reaction
- SAG:
-
senescence associated gene
- X-GAL:
-
5-bromo-4-chloro-3-indolyl-B-d-galactoside
References
Asch F, Wopereis MCS (2001) Responses of field-grown irrigated rice cultivars to varying levels of floodwater salinity in a semi-arid environment. Field Crops Res 70:127–137 doi:10.1016/S0378-4290(01)00128-9
Chai TY, Didierjean L, Burkard G, Genot G (1998) Expression of a green tissue-specific 11 kDa proline-rich protein gene in bean in response to heavy metals. Plant Sci 133:47–56 doi:10.1016/S0168-9452(98)00028-4
Dangel J, Dietrich R, Thomas H (2000) Senescence and programmed cell death. In: Buchanan B, Gruissem W, Jones R (eds) Biochemistry and molecular biology of plants. American Society of Plant Physiologists, Rockville, MD, pp 1044–1100
Dat J, Vandenabeele S, Vranova E, Van Montagu M, Inze D, Van Breusegem F (2000) Dual action of the active oxygen species during plant stress responses. Cell Mol Life Sci 57:779–795 doi:10.1007/s000180050041
Ehrnsperger M, Graber S, Gaestel M, Buchner J (1997) Binding of non-native protein to HSP25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J 16:221–229 doi:10.1093/emboj/16.2.221
Fukuda A, Chiba K (2004) Effect of salt and osmotic stresses on the expression of genes for the vacuolar H+-pyrophosphatase, H+-ATPase subunit A, and Na+ /H+ antiporter from barley. J Exp Bot 55:585–594 doi:10.1093/jxb/erh070
Goodwin W, Pallas JA, Jenkins GI (1996) Transcripts of a gene encoding a putative cell wall-plasma membrane linker protein are specifically cold-induced in Brassica napus. Plant Mol Biol 31:771–781 doi:10.1007/BF00019465
Guterman A, Hjouj AT, Gepstein SH (2003) Senescence-associated mRNAs that may participate in signal transduction and protein trafficking. Physiol Plant 118:439 doi:10.1034/j.1399-3054.2003.00116.x
Hajouj T, Michelis R (2000) Cloning and characterization of a receptor-like protein kinase gene associated with senescence. Plant Physiol 124:1305–1314 doi:10.1104/pp.124.3.1305
He CY, Zhang JS, Chen SY (2002) A soybean gene encoding a proline-rich protein is regulated by salicylic acid, an endogenous circadian rhythm and by various stress. Theor Appl Genet 104:1125–1131 doi:10.1007/s00122-001-0853-5
Hunt R (1978) Plant growth analysis. Edward Arnold, London
Jimenez ES, Medrano L, Martinez-Barajas E (1995) Rubisco activase, a possible new member of the molecular chaperone family. Biochemistry 34:2826–2831 doi:10.1021/bi00009a012
Jose-Estanyol M, Ruiz-Avila L, Puigdomenech P (1992) A maize embryo-specific gene encodes a proline-rich and hydrophobic protein. Plant Cell 4:413–423
Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, Kawai K et al (2001) Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 13:889–905
Khan MSA, Hamid A (1997) Effect of sodium chloride on growth, photosynthesis and mineral ions accumulation of different types of rice (Oryza sativa L.). J Agron Crop Sci 179:149–161 doi:10.1111/j.1439-037X.1997.tb00511.x
Keller B (1993) Structural cell wall proteins. Plant Physiol 101:1127–1130
Koyama ML, Levesley A, Koebner RMD, Flowers TJ, Yeo AR (2001) Quantitative trait loci for component physiological traits determining salt tolerance in rice. Plant Physiol 125:406–422 doi:10.1104/pp.125.1.406
Lamport DTA, Kieliszewski MJ, Showalter AM (2006) Salt stress upregulates periplasmic arabinogalactan proteins: using salt stress to analyze AGP function. New Phytol 169:479–492 doi:10.1111/j.1469-8137.2005.01591.x
Malone M (1992) Kinetics of wound-induced hydraulic signals and variation potentials in wheat seedlings. Planta 187:505–550 doi:10.1007/BF00199969
Menezes-Benavente L, Teixeira FK, Kamei CLA, Margis-Pinheiro M (2004) Salt stress induces altered expression of genes encoding antioxidant enzymes in seedlings of a Brazilian indica rice (Oryza sativa L.). Plant Sci 166:323–331 doi:10.1016/j.plantsci.2003.10.001
Munns R (2002) Comparative physiology of salt and water stress. Plant Cell Environ 25:239–250 doi:10.1046/j.0016-8025.2001.00808.x
Munns R, Schachtman DP, Condon AG (1995) The significant of a two-phase growth response to salinity in wheat and barley. Aust J Plant Physiol 22:561–569
Navabpour S, Morris K (2003) Expression of senescence-enhanced genes in response to oxidative stress. J Exp Bot 54:2285–2292 doi:10.1093/jxb/erg267
Neumann PM (1997) Salinity resistance and plant growth revisited. Plant Cell Environ 20:1193–1198 doi:10.1046/j.1365-3040.1997.d01-139.x
Pillai MA, Yanagihara S (2001) Molecular cloning and characterization of salt responsive genes in rice (Oryza sativa). J Plant Physiol 158:1189–1194 doi:10.1078/0176-1617-00247
Pont-Lezica RF, McNally JG, Pickard BG (1993) Wall-to-membrane linkers in onion epidermis: some hypotheses. Plant Cell Environ 16:111–123 doi:10.1111/j.1365-3040.1993.tb00853.x
Ristic Z, Ashworth EN (1994) Response of xylem ray parenchyma cells of red oiser dogwood (Cornus sericea L.) to freezing stress. Plant Physiol 104:737–746
Saneoka H, Ishiguro S (2001) Effect of salinity and abscisic acid on accumulation of glycinebetaine and betaine aldehyde dehydrogenase mRNA in Sorghum leaves (Sorghum bicolor). J Plant Physiol 158:853–859 doi:10.1078/0176-1617-00058
Sheng J, D'Ovidio R, Mehdy MC (1991) Negative and positive regulation of a novel proline-rich protein mRNA by fungal elicitor and wounding. Plant J 1:345–354 doi:10.1046/j.1365-313X.1991.t01-3-00999.x
Sun W (2002) Small heat shock proteins and stress tolerance in plants. Biochim Biophys Acta 1577:1–9
Thompson JE, Froese CD (1998) Lipid metabolism during plant senescence. Prog Lipid Res 37:119–141 doi:10.1016/S0163-7827(98)00006-X
Torok Z, Goloubinoff P, Horvath I, Tsvetkova NM, Glatz A, Balogh G et al (2001) Synechocystis HSP17 is an amphitropic protein that stabilizes heat-stressed membranes and binds denatured proteins for subsequent chaperone-mediated refolding. Proc Natl Acad Sci U S A 98:3098–3103 doi:10.1073/pnas.051619498
Umezawa T, Mizuno K, Fujimura T (2002) Discrimination of genes expressed in response to the ionic or osmotic effect of salt stress in soybean with cDNA-AFLP. Plant Cell Environ 25:1617–1625 doi:10.1046/j.1365-3040.2002.00939.x
Vaidyanathan H, Sivakumar P, Chakrabarty R, Thomas G (2003) Scavenging of reactive oxygen species in NaCl-stressed rice (Oryza sativa L.) – differential response in salt-tolerant and sensitive varieties. Plant Sci 165:1411–1418 doi:10.1016/j.plantsci.2003.08.005
Verdoy D, Lucas MM, Manrique E, Covarrubias A, De Filipe MR, Pueyo JJ (2004) Differential organ-specific response to salt stress and water deficit in nodulated bean (Phaseolus vulgaris). Plant Cell Environ 27:757–767 doi:10.1111/j.1365-3040.2004.01179.x
Vos P, Hogers R, Bleeker M, Rejian M, Lee TVD, Hormes M et al (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414 doi:10.1093/nar/23.21.4407
Wang WX, Vincour B, Altman A (2003) Plant responses to drought, salinity and extreme temperatures; towards genetic engineering for stress tolerance. Planta 218:1–14 doi:10.1007/s00425-003-1105-5
Yeo AR (1992) Variation and inheritance of sodium transport in rice. Plant Soil 146:109–116 doi:10.1007/BF00012002
Yeo AR, Lee KS, Izard P, Boursier PJ, Flowers TJ (1991) Short-term and long-term effects of salinity on leaf growth in rice (Oryza-Sativa L). J Exp Bot 42:881–889 doi:10.1093/jxb/42.7.881
Yokoi S, Quintero FJ, Cubero Ruiz MT, Bressan RA, Hasegawa PM, Pardo JM (2002) Differential expression and function of Arabidopsis thaliana NHX, Na+ /H+ antiporters in the salt stress response. Plant J 30:529–539 doi:10.1046/j.1365-313X.2002.01309.x
Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Laboratory manual for physiological studies of rice, 3rd edn. International Rice Research Institute, Manila
Yu RMK, Zhou Y (2003) Two genes encoding protein phosphatase 2A catalytic subunits are differentially expressed in rice. Plant Mol Biol 51:295–311
Acknowledgment
The authors wish to thank Dr. Mike Malone for his help and encouragement. These experiments were conducted at the School of Life Science of the University of Sussex, Brighton, UK and supported by a scholarship from the Ministry of Sciences, Research and Biotechnology, Iran.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: John McPherson Cheeseman.
Rights and permissions
About this article
Cite this article
Roshandel, P., Flowers, T. The ionic effects of NaCl on physiology and gene expression in rice genotypes differing in salt tolerance. Plant Soil 315, 135–147 (2009). https://doi.org/10.1007/s11104-008-9738-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-008-9738-6