Abstract
Key message
A transcriptome analysis reveals the transcripts and alleles differentially expressed in sugarcane genotypes with contrasting lignin composition.
Abstract
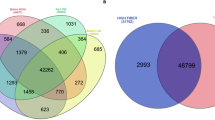
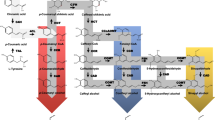
Sugarcane bagasse is a highly abundant resource that may be used as a feedstock for the production of biofuels and bioproducts in order to meet increasing demands for renewable replacements for fossil carbon. However, lignin imparts rigidity to the cell wall that impedes the efficient breakdown of the biomass into fermentable sugars. Altering the ratio of the lignin units, syringyl (S) and guaiacyl (G), which comprise the native lignin polymer in sugarcane, may facilitate the processing of bagasse. This study aimed to identify genes and markers associated with S/G ratio in order to accelerate the development of sugarcane bioenergy varieties with modified lignin composition. The transcriptome sequences of 12 sugarcane genotypes that contrasted for S/G ratio were compared and there were 2019 transcripts identified as differentially expressed (DE) between the high and low S/G ratio groups. These included transcripts encoding possible monolignol biosynthetic pathway enzymes, transporters, dirigent proteins and transcriptional and post-translational regulators. Furthermore, the frequencies of single nucleotide polymorphisms (SNPs) were compared between the low and high S/G ratio groups to identify specific alleles expressed with the phenotype. There were 2063 SNP loci across 787 unique transcripts that showed group-specific expression. Overall, the DE transcripts and SNP alleles identified in this study may be valuable for breeding sugarcane varieties with altered S/G ratio that may provide desirable bioenergy traits.




Similar content being viewed by others
Data Availability
All raw RNA-seq read data is available from the NCBI SRA database under the BioProject PRJNA628577, https://www.ncbi.nlm.nih.gov/bioproject/628577.
References
Aden A, Ruth M, Ibsen K, Jechura J, Neeves K, Sheehan J, Wallace B, Montague L, Slayton A, Lukas J (2002) Lignocellulosic biomass to ethanol process design and economics utilizing co-current dilute acid prehydrolysis and enzymatic hydrolysis for corn stover. NREL Lab Anal. Proced. (LAP), Tech. report, NREL/TP-510–32438.
Alejandro S, Lee Y, Tohge T, Sudre D, Osorio S, Park J, Bovet L, Lee Y, Geldner N, Fernie AR, Martinoia E (2012) AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr Biol 22:1207–1212
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Barakat A, Yassin NBM, Park JS, Choi A, Herr J, Carlson JE (2011) Comparative and phylogenomic analyses of cinnamoyl-CoA reductase and cinnamoyl-CoA-reductase-like gene family in land plants. Plant Sci 181:249–257
Barrière Y, Courtial A, Chateigner-Boutin AL, Denoue D, Grima-Pettenati J (2015) Breeding maize for silage and biofuel production, an illustration of a step forward with the genome sequence. Plant Sci 242:310–329
Baucher M, Bernard-Vailhé MA, Chabbert B, Besle JM, Opsomer C, Van Montagu M, Botterman J (1999) Down-regulation of cinnamyl alcohol dehydrogenase in transgenic alfalfa (Medicago sativa L.) and the effect on lignin composition and digestibility. Plant Mol Biol 39:437–447
Benjamin Y, Cheng H, Görgens JF (2013) Evaluation of bagasse from different varieties of sugarcane by dilute acid pretreatment and enzymatic hydrolysis. Ind Crops Prod 51:7–18
Bertrand E, Vandenberghe LP, Soccol CR, Sigoillot J-C, Faulds C (2016) First generation bioethanol. In: Soccol CR, Brar SK, Faulds C, Ramos LP (eds) Green fuels technology, green energy and technology. Springer International Publishing, Switzerland, pp 175–212
Bewg WP, Poovaiah C, Lan W, Ralph J, Coleman HD (2016) RNAi downregulation of three key lignin genes in sugarcane improves glucose release without reduction in sugar production. Biotechnol Biofuels 9:1–13
Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54:519–546. https://doi.org/10.1146/annurev.arplant.54.031902.134938
Bonawitz ND, Kim JI, Tobimatsu Y, Ciesielski PN, Anderson NA, Ximenes E, Maeda J, Ralph J, Donohoe BS, Ladisch M, Chapple C (2014) Disruption of mediator rescues the stunted growth of a lignin-deficient arabidopsis mutant. Nature 509:376–380. https://doi.org/10.1038/nature13084
Borah P, Khurana JP (2018) The OsFBK1 E3 ligase subunit affects anther and root secondary cell wall thickenings by mediating turnover of a cinnamoyl-CoA reductase. Plant Physiol 176:2148–2165. https://doi.org/10.1104/pp.17.01733
Bosch M, Mayer CD, Cookson A, Donnison IS (2011) Identification of genes involved in cell wall biogenesis in grasses by differential gene expression profiling of elongating and non-elongating maize internodes. J Exp Bot 62:3545–3561. https://doi.org/10.1093/jxb/err045
Bottcher A, Cesarino I, Brombini dos Santos A, Vicentini R, Mayer JLS, Vanholme R, Morreel K, Goeminne G, Moura JCMS, Nobile PM, Carmello-Guerreiro SM, Antonio dos Anjos I, Creste S, Boerjan W, Landell MG (2013) Lignification in sugarcane: biochemical characterization, gene discovery, and expression analysis in two genotypes contrasting for lignin content. Plant Physiol 163:1539–1557. https://doi.org/10.1104/pp.113.225250
Cao Y, Han Y, Meng D, Abdullah M, Li D, Jin Q, Lin Y, Cai Y (2018) Systematic analysis and comparison of the PHD-finger gene family in Chinese pear (Pyrus bretschneideri) and its role in fruit development. Funct Integr Genomics 18:519–531. https://doi.org/10.1007/s10142-018-0609-9
Cassan-Wang H, Goué N, Saidi MN, Legay S, Sivadon P, Goffner D, Grima-Pettenati J (2013) Identification of novel transcription factors regulating secondary cell wall formation in Arabidopsis. Front Plant Sci 4:1–14. https://doi.org/10.3389/fpls.2013.00189
Chao N, Li N, Qi Q, Li S, Lv T, Jiang XN, Gai Y (2017) Characterization of the cinnamoyl-CoA reductase (CCR) gene family in Populus tomentosa reveals the enzymatic active sites and evolution of CCR. Planta 245:61–75. https://doi.org/10.1007/s00425-016-2591-6
Conesa A, Götz S (2008) Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 2008:619832. https://doi.org/10.1155/2008/619832
Damaj MB, Kumpatla SP, Emani C, Beremand PD, Reddy AS, Rathore KS, Buenrostro-Nava MT, Curtis IS, Thomas TL, Mirkov EE (2010) Sugarcane dirigent and O-methyltransferase promoters confer stem-regulated gene expression in diverse monocots. Planta 231:1439–1458. https://doi.org/10.1007/s00425-010-1138-5
Davison BH, Drescher SR, Tuskan GA, Davis MF, Nghiem NP (2006) Variation of S/G ratio and lignin content in a Populus family influences the release of xylose by dilute acid hydrolysis. Appl Biochem Biotechnol 130:427–435. https://doi.org/10.1385/ABAB:130:1:427
Del Río JC, Prinsen P, Rencoret J, Nieto L, Jiménez-Barbero J, Ralph J, Martínez ÁT, Gutiérrez A (2012) Structural characterization of the lignin in the cortex and pith of elephant grass (Pennisetum purpureum) stems. J Agric Food Chem 60:3619–3634. https://doi.org/10.1021/jf300099g
del Río JC, Lino AG, Colodette JL, Lima CF, Gutiérrez A, Martínez ÁT, Lu F, Ralph J, Rencoret J (2015) Differences in the chemical structure of the lignins from sugarcane bagasse and straw. Biomass Bioenerg 81:322–338. https://doi.org/10.1016/j.biombioe.2015.07.006
Ehlting È, Bu D, Wang Q, Kombrink E (1999) Three 4-coumarate;coenzyme a ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J 19:9–20
Eudes A, Dutta T, Deng K, Jacquet N, Sinha A, Benites VT, Baidoo EEK, Riche A, Sattler SE, Northen TR, Singh S, Simmons BA, Loqué D (2017) SbCOMT (Bmr12) is involved in the biosynthesis of tricin-lignin in sorghum. PLoS ONE 12:1–11. https://doi.org/10.1371/journal.pone.0178160
Fan D, Li C, Fan C, Hu J, Li J, Yao S, Lu W, Yan Y, Luo K (2020) MicroRNA6443-mediated regulation of FERULATE 5-HYDROXYLASE gene alters lignin composition and enhances saccharification in Populus tomentosa. New Phytol 226:410–425. https://doi.org/10.1111/nph.16379
FAOSTAT (2018) Food and Agriculture Organization of the United Nations
Fornalé S, Capellades M, Encina A, Wang K, Irar S, Lapierre C, Ruel K, Joseleau JP, Berenguer J, Puigdomènech P, Rigau J, Caparrós-Ruiz D (2012) Altered lignin biosynthesis improves cellulosic bioethanol production in transgenic maize plants down-regulated for cinnamyl alcohol dehydrogenase. Mol Plant 5:817–830. https://doi.org/10.1093/mp/ssr097
Fornalé S, Rencoret J, García-Calvo L, Encina A, Rigau J, Gutiérrez A, Del Río JC, Caparros-Ruiz D (2017) Changes in cell wall polymers and degradability in maize mutants lacking 30- and 50-o-methyltransferases involved in lignin biosynthesis. Plant Cell Physiol 58:240–255. https://doi.org/10.1093/pcp/pcw198
Franke R, McMichael CM, Meyer K, Shirley AM, Cusumano JC, Chappie C (2000) Modified lignin in tobacco and poplar plants over-expressing the Arabidopsis gene encoding ferulate 5-hydroxylase. Plant J 22:223–234. https://doi.org/10.1046/j.1365-313X.2000.00727.x
Fu C, Mielenz JR, Xiao X, Ge Y, Hamilton CY, Rodriguez M, Chen F, Foston M, Ragauskasc A, Bouton J, Dixon RA, Wang Z-Y (2011) Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc Natl Acad Sci 108:3803–3808. https://doi.org/10.1073/pnas.1100310108
Fu Y, Win P, Zhang H, Li C, Shen Y, He F, Luo K (2019) PtrARF2.1 is involved in regulation of leaf development and lignin biosynthesis in poplar trees. Int J Mol Sci 20:4141. https://doi.org/10.3390/ijms20174141
Funatsuki H, Suzuki M, Hirose A, Inaba H, Yamada T, Hajika M, Komatsu K, Katayama T, Sayama T, Ishimoto M, Fujino K (2014) Molecular basis of a shattering resistance boosting global dissemination of soybean. Proc Natl Acad Sci 111:17797–17802. https://doi.org/10.1073/pnas.1417282111
Furtado A (2014) DNA extraction from vegetative tissue for next-generation sequencing. In: Henry RJ, Furtado A (eds) Cereal Genomics: Methods and Protocols. Springer Science+Business Media, New York, NY, USA, pp 1–5
Goujon T, Sibout R, Pollet B, Maba B, Nussaume L, Bechtold N, Lu F, Ralph J, Mila I, Barrière Y, Lapierre C, Jouanin L (2003) A new Arabidopsis thaliana mutant deficient in the expression of O-methyltransferase impacts lignins and sinapoyl esters. Plant Mol Biol 51:973–989. https://doi.org/10.1023/A:1023022825098
Grabber JH (2005) How do lignin composition, structure, and cross-linking affect degradability? A review of cell wall model studies. Crop Sci 45:820–831. https://doi.org/10.2135/cropsci2004.0191
Guo D, Chen F, Inoue K, Blount JW, Dixon RA (2001) Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa: impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13:73–88. https://doi.org/10.1105/tpc.13.1.73
Han LB, Li YB, Wang HY, Wu XM, Li CL, Luo M, Wu SJ, Kong ZS, Pei Y, Jiao GL, Xia GX (2013) The dual functions of WLIM1a in cell elongation and secondary wall formation in developing cotton fibers. Plant Cell 25:4421–4438. https://doi.org/10.1105/tpc.113.116970
Hoang NV, Furtado A, Mason PJ, Marquardt A, Kasirajan L, Thirugnanasambandam PP, Botha FC, Henry RJ (2017a) A survey of the complex transcriptome from the highly polyploid sugarcane genome using full-length isoform sequencing and de novo assembly from short read sequencing. BMC Genomics 18:395. https://doi.org/10.1186/s12864-017-3757-8
Hoang NV, Furtado A, O’Keeffe AJ, Botha FC, Henry RJ (2017b) Association of gene expression with biomass content and composition in sugarcane. PLoS ONE 12:e0183417. https://doi.org/10.1371/journal.pone.0183417
Hoang NV, Furtado A, Donnan L, Keeffe EC, Botha FC, Henry RJ (2017c) High-throughput profiling of the fiber and sugar composition of sugarcane biomass. Bioenergy Res 10:400–416. https://doi.org/10.1007/s12155-016-9801-8
Hodgson-Kratky K, Papa G, Rodriguez A, Stavila V, Simmons B, Botha F, Furtado A, Henry R (2019) Relationship between sugarcane culm and leaf biomass composition and saccharification efficiency. Biotechnol Biofuels 12:247. https://doi.org/10.1186/s13068-019-1588-3
Hosmani PS, Kamiya T, Danku J, Naseer S, Geldner N, Guerinot ML, Salt DE (2013) Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. Proc Natl Acad Sci 110:14498–14503. https://doi.org/10.1073/pnas.1308412110
Ho-Yue-Kuang S, Alvarado C, Antelme S, Bouchet B, Cézard L, Le Bris P, Legée F, Maia-Grondard A, Yoshinaga A, Saulnier L, Guillon F, Sibout R, Lapierre C, Chateigner-Boutin AL (2016) Mutation in Brachypodium caffeic acid O-methyltransferase 6 alters stem and grain lignins and improves straw saccharification without deteriorating grain quality. J Exp Bot 67:227–237. https://doi.org/10.1093/jxb/erv446
Hu R, Xu Y, Yu C, He K, Tang Q, Jia C, He G, Wang X, Kong Y, Zhou G (2017) Transcriptome analysis of genes involved in secondary cell wall biosynthesis in developing internodes of Miscanthus lutarioriparius. Sci Rep 7:1–16. https://doi.org/10.1038/s41598-017-08690-8
Humphreys JM, Hemm MR, Chapple C (1999) New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc Natl Acad Sci U S A 96:10045–10050. https://doi.org/10.1073/pnas.96.18.10045
Jun SY, Walker AM, Kim H, Ralph J, Vermerris W, Sattler SE, Kang CH (2017) The enzyme activity and substrate specificity of two major cinnamyl alcohol dehydrogenases in Sorghum (Sorghum bicolor), SbCAD2 and SbCAD4. Plant Physiol 174:2128–2145. https://doi.org/10.1104/pp.17.00576
Jun SY, Sattler SA, Cortez GS, Vermerris W, Sattler SE, Kang CH (2018) Biochemical and structural analysis of substrate specificity of a phenylalanine ammonia-lyase. Plant Physiol 176:1452–1468. https://doi.org/10.1104/pp.17.01608
Jung JH, Altpeter F (2016) TALEN mediated targeted mutagenesis of the caffeic acid O-methyltransferase in highly polyploid sugarcane improves cell wall composition for production of bioethanol. Plant Mol Biol 92:131–142. https://doi.org/10.1007/s11103-016-0499-y
Jung JH, Fouad WM, Vermerris W, Gallo M, Altpeter F (2012) RNAi suppression of lignin biosynthesis in sugarcane reduces recalcitrance for biofuel production from lignocellulosic biomass. Plant Biotechnol J 10:1067–1076. https://doi.org/10.1111/j.1467-7652.2012.00734.x
Jung JH, Vermerris W, Gallo M, Fedenko JR, Erickson JE, Altpeter F (2013) RNA interference suppression of lignin biosynthesis increases fermentable sugar yields for biofuel production from field-grown sugarcane. Plant Biotechnol J 11:709–716. https://doi.org/10.1111/pbi.12061
Jung JH, Kannan B, Dermawan H, Moxley GW, Altpeter F (2016) Precision breeding for RNAi suppression of a major 4-coumarate:coenzyme A ligase gene improves cell wall saccharification from field grown sugarcane. Plant Mol Biol 92:505–517. https://doi.org/10.1007/s11103-016-0527-y
Kajita S, Hishiyama S, Tomimura Y, Katayama Y, Omori S (1997) Structural characterization of modified lignin in transgenic tobacco plants in which the activity of 4-coumarate:coenzyme A ligase is depressed. Plant Physiol 114:871–879. https://doi.org/10.1104/pp.114.3.871
Kannan B, Jung JH, Moxley GW, Lee SM, Altpeter F (2018) TALEN-mediated targeted mutagenesis of more than 100 COMT copies/alleles in highly polyploid sugarcane improves saccharification efficiency without compromising biomass yield. Plant Biotechnol J 16:856–866. https://doi.org/10.1111/pbi.12833
Kasirajan L, Hoang NV, Furtado A, Botha FC, Henry RJ (2018) Transcriptome analysis highlights key differentially expressed genes involved in cellulose and lignin biosynthesis of sugarcane genotypes varying in fiber content. Sci Rep 8:11612. https://doi.org/10.1038/s41598-018-30033-4
Kawaoka A, Kaothien P, Yoshida K, Endo S, Yamada K, Ebinuma H (2000) Functional analysis of tobacco LIM protein Ntlim1 involved in lignin biosynthesis. Plant J 22:289–301. https://doi.org/10.1046/j.1365-313X.2000.00737.x
Kawaoka A, Nanto K, Ishii K, Ebinuma H (2006) Reduction of lignin content by suppression of expression of the LIM domain transcription factor in Eucalyptus camaldulensis. Silvae Genet 55:269–277. https://doi.org/10.1515/sg-2006-0035
Khanna M, Hochman G, Rajagopal D, Sexton S, Zilberman D (2009) Sustainability of food, energy and environment with biofuels. CAB Rev Perspect Agric Vet Sci Nutr Nat Resour 4:1–10. https://doi.org/10.1079/PAVSNNR20094028
Kishimoto T, Chiba W, Saito K, Fukushima K, Uraki Y, Ubukata M (2010) Influence of syringyl to guaiacyl ratio on the structure of natural and synthetic lignins. J Agric Food Chem 58:895–901. https://doi.org/10.1021/jf9035172
Ko JH, Kim WC, Han KH (2009) Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. Plant J 60:649–665. https://doi.org/10.1111/j.1365-313X.2009.03989.x
Kumar A, Ellis BE (2003) 4-Coumarate:CoA ligase gene family in Rubus idaeus: cDNA structures, evolution, and expression. Plant Mol Biol 51:327–340. https://doi.org/10.1023/A:1022004923982
Lacombe E, Hawkins S, Van Doorsselaere J, Piquemal J, Goffner D, Poeydomenge O, Boudet AM, Grima-Pettenati J (1997) Cinnamoyl CoA reductase, the first committed enzyme of the lignin branch biosynthetic pathway: cloning, expression and phylogenetic relationships. Plant J 11:429–441. https://doi.org/10.1046/j.1365-313X.1997.11030429.x
Lapierre C, Pollet B, Petit-Conil M, Toval G, Romero J, Pilate G, Leplé J-C, Boerjan W, Ferret V, De Nadai V, Jouanin L (1999) Structural alterations of lignins in transgenic poplars with depressed cinnamyl alcohol dehydrogenase or caffeic acid O-methyltransferase activity have an opposite impact on the efficiency of industrial kraft pulping. Plant Physiol 119:153–164. https://doi.org/10.1104/pp.119.1.153
Lauvergeat V, Lacomme C, Lacombe E, Lasserre E, Roby D, Grima-Pettenati J (2001) Two cinnamoyl-CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry 57:1187–1195. https://doi.org/10.1016/S0031-9422(01)00053-X
Law CW, Chen Y, Shi W, Smyth GK (2014) Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15:R29. https://doi.org/10.1186/gb-2014-15-2-r29
Law CW, Alhamdoosh M, Su S, Dong X, Tian L, Smyth GK, Ritchie ME (2016) RNA-seq analysis is easy as 1–2–3 with limma, Glimma and edgeR F1000Research 5:ISCB. Comm J. https://doi.org/10.12688/F1000RESEARCH.9005.1
Leplé JC, Dauwe R, Morreel K, Storme V, Lapierre C, Pollet B, Naumann A, Kang KY, Kim H, Ruel K, Lefèbvre A, Joseleau JP, Grima-Pettenati J, De Rycke R, Andersson-Gunnerås S, Erban A, Fehrle I, Petit-Conil M, Kopka J, Polle A, Messens E, Sundberg B, Mansfield SD, Ralph J, Pilate G, Boerjan W (2007) Downregulation of cinnamoyl-coenzyme a reductase in poplar: multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell 19:3669–3691. https://doi.org/10.1105/tpc.107.054148
Li L, Popko JL, Umezawa T, Chiang VL (2000) 5-Hydroxyconiferyl aldehyde modulates enzymatic methylation for syringyl monolignol formation, a new view of monolignol biosynthesis in angiosperms. J Biol Chem 275:6537–6545. https://doi.org/10.1074/jbc.275.9.6537
Li X, Ximenes E, Kim Y, Slininger M, Meilan R, Ladisch M, Chapple C (2010) Lignin monomer composition affects Arabidopsis cell-wall degradability after liquid hot water pretreatment. Biotechnol Biofuels 3:1–7. https://doi.org/10.1186/1754-6834-3-27
Li X, Chen W, Zhao Y, Xiang Y, Jiang H, Zhu S, Cheng B (2013) Downregulation of caffeoyl-CoA O-methyltransferase (CCoAOMT) by RNA interference leads to reduced lignin production in maize straw. Genet Mol Biol 36:540–546. https://doi.org/10.1590/S1415-47572013005000039
Li N, Lin B, Wang H, Li X, Yang F, Ding X, Yan J, Chu Z (2019) Natural variation in Zm FBL41 confers banded leaf and sheath blight resistance in maize. Nat Genet 51:1540–1548. https://doi.org/10.1038/s41588-019-0503-y
Lu S, Li Q, Wei H, Chang M-J, Tunlaya-Anukit S, Kim H, Liu J, Song J, Sun Y-H, Yuan L, Yeh T-F, Peszlen I, Ralph J, Sederoff RR, Chiang VL (2013) Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc Natl Acad Sci 110:10848–10853. https://doi.org/10.1073/pnas.1308936110
Malik S, Roeder RG (2010) The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet 11:761–772. https://doi.org/10.1038/nrg2901
Martins APB, dos Brito M, Mayer S, Llerena JLS, Oliveira JPP, Takahashi JF, Carlin NG, Borges SD, Andrade DNAF, Peixoto-Júnior LM, Goldman RF, Mazzafera S, Creste P, Nobile S (2018) Ectopic expression of sugarcane SHINE changes cell wall and improves biomass in rice. Biomass Bioenerg 119:322–334. https://doi.org/10.1016/j.biombioe.2018.09.036
Masarin F, Gurpilhares DB, Baffa DC, Barbosa MH, Carvalho W, Ferraz A, Milagres AM (2011) Chemical composition and enzymatic digestibility of sugarcane clones selected for varied lignin content. Biotechnol Biofuels. https://doi.org/10.1186/1754-6834-4-55
Mewalal R, Mizrachi E, Coetzee B, Mansfield SD, Myburg AA (2016) The Arabidopsis domain of unknown function 1218 (DUF1218) containing proteins, MODIFYING WALL LIGNIN-1 and 2 (At1g31720/MWL-1 and At4g19370/MWL-2) function redundantly to alter secondary cell wall lignin content. PLoS ONE 11:e0150254. https://doi.org/10.1371/journal.pone.0150254
Murciano Martínez P, Punt AM, Kabel MA, Gruppen H (2016) Deconstruction of lignin linked p-coumarates, ferulates and xylan by NaOH enhances the enzymatic conversion of glucan. Bioresour Technol 216:44–51. https://doi.org/10.1016/j.biortech.2016.05.040
Nobile PM, Bottcher A, Mayer JLS, Brito MS, dos Anjos IA, Landell MGA, Vicentini R, Creste S, Riaño-Pachón DM, Mazzafera P (2017) Identification, classification and transcriptional profiles of dirigent domain-containing proteins in sugarcane. Mol Genet Genomics 292:1323–1340. https://doi.org/10.1007/s00438-017-1349-6
Noguchi M, Fujiwara M, Sano R, Nakano Y, Fukao Y, Ohtani M, Demura T (2018) Proteomic analysis of xylem vessel cell differentiation in VND7-inducible tobacco BY-2 cells by two-dimensional gel electrophoresis. Plant Biotechnol 35:31–37. https://doi.org/10.5511/plantbiotechnology.18.0129a
Osakabe K, Tsao CC, Li L, Popko JL, Umezawa T, Carraway DT, Smeltzer RH, Joshi CP, Chiang VL (1999) Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proc Natl Acad Sci U S A 96:8955–8960. https://doi.org/10.1073/pnas.96.16.8955
Papa G, Varanasi P, Sun L, Cheng G, Stavila V, Holmes B, Simmons BA, Adani F, Singh S (2012) Exploring the effect of different plant lignin content and composition on ionic liquid pretreatment efficiency and enzymatic saccharification of Eucalyptus globulus L. mutants. Bioresour Technol 117:352–359. https://doi.org/10.1016/j.biortech.2012.04.065
Park JJ, Yoo CG, Flanagan A, Pu Y, Debnath S, Ge Y, Ragauskas AJ, Wang ZY (2017) Defined tetra-allelic gene disruption of the 4-coumarate:coenzyme A ligase 1 (Pv4CL1) gene by CRISPR/Cas9 in switchgrass results in lignin reduction and improved sugar release. Biotechnol Biofuels 10:284. https://doi.org/10.1186/s13068-017-0972-0
Peng X, Qin Z, Zhang G, Guo Y, Huang J (2015) Integration of the proteome and transcriptome reveals multiple levels of gene regulation in the rice dl2 mutant. Front Plant Sci 6:351. https://doi.org/10.3389/fpls.2015.00351
Penning BW, Sykes RW, Babcock NC, Dugard CK, Held MA, Klimek JF, Shreve JT, Fowler M, Ziebell A, Davis MF, Decker SR, Turner GB, Mosier NS, Springer NM, Thimmapuram J, Weil CF, Mccann MC, Carpita NC (2014) Genetic determinants for enzymatic digestion of lignocellulosic biomass are independent of those for lignin abundance in a maize recombinant inbred population. Plant Physiol 165:1475–1487. https://doi.org/10.1104/pp.114.242446
Petzold HE, Rigoulot SB, Zhao C, Chanda B, Sheng X, Zhao M, Jia X, Dickerman AW, Beers EP, Brunner AM (2017) Identification of new protein-protein and protein-DNA interactions linked with wood formation in Populus trichocarpa. Tree Physiol 38:362–377. https://doi.org/10.1093/treephys/tpx121
Poovaiah CR, Bewg WP, Lan W, Ralph J, Coleman HD (2016) Sugarcane transgenics expressing MYB transcription factors show improved glucose release. Biotechnol Biofuels 9:143. https://doi.org/10.1186/s13068-016-0559-1
Ralph J, Hatfield RD (1991) Pyrolysis-GC-MS characterization of forage materials. J Agric Food Chem 39:1426–1437. https://doi.org/10.1021/jf00008a014
Reddy MSS, Chen F, Shadle G, Jackson L, Aljoe H, Dixon RA (2005) Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.). Proc Natl Acad Sci U S A 102:16573–16578. https://doi.org/10.1073/pnas.0505749102
Riboulet C, Guillaumie S, Méchin V, Bosio M, Pichon M, Goffner D, Lapierre C, Pollet B, Lefevre B, Martinant JP, Barrière Y (2009) Kinetics of phenylpropanoid gene expression in maize growing internodes: relationships with cell wall deposition. Crop Sci 49:211–223. https://doi.org/10.2135/cropsci2008.03.0130
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. https://doi.org/10.1093/nar/gkv007
Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11:R25. https://doi.org/10.1186/gb-2010-11-3-r25
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. https://doi.org/10.1093/bioinformatics/btp616
Saballos A, Ejeta G, Sanchez E, Kang C, Vermerris W (2009) A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum [Sorghum bicolor (L.) Moench] identifies SbCAD2 as the brown midrib6 gene. Genetics 181:783–795. https://doi.org/10.1534/genetics.108.098996
Saballos A, Sattler SE, Sanchez E, Foster TP, Xin Z, Kang C, Pedersen JF, Vermerris W (2012) Brown midrib2 (Bmr2) encodes the major 4-coumarate: Coenzyme A ligase involved in lignin biosynthesis in sorghum (Sorghum bicolor (L.) Moench). Plant J 70:818–830. https://doi.org/10.1111/j.1365-313X.2012.04933.x
Sánchez-Elordi E, Contreras R, de Armas R, Benito MC, Alarcón B, de Oliveira E, del Mazo C, Díaz-Peña EM, Santiago R, Vicente C, Legaz ME (2018) Differential expression of SofDIR16 and SofCAD genes in smut resistant and susceptible sugarcane cultivars in response to Sporisorium scitamineum. J Plant Physiol 226:103–113. https://doi.org/10.1016/j.jplph.2018.04.016
Santos FA, De Queiroz JH, Colodette JL, Manfredi M, Queiroz MELR, Caldas CS, Soares FEF (2014) Otimização do pré-tratamento hidrotérmico da palha de cana-de-açúcar visando à produção de etanol celulósico. Quim Nova 37:56–62. https://doi.org/10.1590/S0100-40422014000100011
Sattler SE, Saathoff AJ, Haas EJ, Palmer NA, Funnell-Harris DL, Sarath G, Pedersen JF (2009) A nonsense mutation in a cinnamyl alcohol dehydrogenase gene is responsible for the sorghum brown midrib6 phenotype. Plant Physiol 150:584–595. https://doi.org/10.1104/pp.109.136408
Schwacke R, Ponce-Soto GY, Krause K, Bolger AM, Arsova B, Hallab A, Gruden K, Stitt M, Bolger ME, Usadel B (2019) MapMan4: a refined protein classification and annotation framework applicable to multi-omics data analysis. Mol Plant 12:879–892. https://doi.org/10.1016/j.molp.2019.01.003
Scully ED, Gries T, Sarath G, Palmer NA, Baird L, Serapiglia MJ, Dien BS, Boateng AA, Ge Z, Funnell-Harris DL, Twigg P, Clemente TE, Sattler SE (2016) Overexpression of SbMyb60 impacts phenylpropanoid biosynthesis and alters secondary cell wall composition in Sorghum bicolor. Plant J 85:378–395. https://doi.org/10.1111/tpj.13112
Selig MJ, Tucker MP, Sykes RW, Reichel KL, Brunecky R, Himmel ME, Davis MF, Decker SR (2010) Lignocellulose recalcitrance screening by integrated high-throughput hydrothermal pretreatment and enzymatic saccharification. Ind Biotechnol 6:104–111. https://doi.org/10.1089/ind.2010.0009
Sewalt VJH, Ni W, Blount JW, Jung HG, Masoud SA, Howles PA, Lamb C, Dixon RA (1997) Reduced lignin content and altered lignin composition in transgenic tobacco down-regulated in expression of L-phenylalanine ammonia-lyase or cinnamate 4-hydroxylase. Plant Physiol 115:41–50. https://doi.org/10.1104/pp.115.1.41
Seyednasrollah F, Laiho A, Elo LL (2013) Comparison of software packages for detecting differential expression in RNA-seq studies. Brief Bioinform 16:59–70. https://doi.org/10.1093/bib/bbt086
Skirycz A, Jozefczuk S, Stobiecki M, Muth D, Zanor MI, Witt I, Mueller-roeber B (2007) Transcription factor AtDOF4;2 affects phenylpropanoid metabolism in Arabidopsis thaliana. New Phytol 175:425–438
Sonbol FM, Fornalé S, Capellades M, Encina A, Touriño S, Torres JL, Rovira P, Ruel K, Puigdomènech P, Rigau J, Caparrós-Ruiz D (2009) The maize ZmMYB42 represses the phenylpropanoid pathway and affects the cell wall structure, composition and degradability in Arabidopsis thaliana. Plant Mol Biol 70:283–296. https://doi.org/10.1007/s11103-009-9473-2
Soneson C, Delorenzi M (2013) A comparison of methods for differential expression analysis of RNA-seq data. BMC Bioinformatics 14:91. https://doi.org/10.1186/1471-2105-14-91
Stare T, Stare K, Weckwerth W, Wienkoop S, Gruden K (2017) Comparison between proteome and transcriptome response in potato (Solanum tuberosum L) leaves following potato virus Y (PVY) infection. Proteomes 5:14. https://doi.org/10.3390/proteomes5030014
Stewart JJ, Akiyama T, Chapple C, Ralph J, Mansfield SD (2009) The effects on lignin structure of overexpression of ferulate 5-hydroxylase in hybrid poplar. Plant Physiol 150:621–635. https://doi.org/10.1104/pp.109.137059
Studer MH, DeMartini JD, Davis MF, Sykes RW, Davison B, Keller M, Tuskan GA, Wyman CE (2011) Lignin content in natural Populus variants affects sugar release. Proc Natl Acad Sci 108:6300–6305. https://doi.org/10.1073/pnas.1009252108
Sundin L, Vanholme R, Geerinck J, Goeminne G, Höfer R, Kim H, Ralph J, Boerjan W (2014) Mutation of the inducible arabidopsis thaliana cytochrome P450 reductase2 alters lignin composition and improves saccharification. Plant Physiol 166:1956–1971. https://doi.org/10.1104/pp.114.245548
Sykes RW, Gjersing EL, Foutz K, Rottmann WH, Kuhn SA, Foster CE, Ziebell A, Turner GB, Decker SR, Hinchee MAW, Davis MF (2015) Down-regulation of p-coumaroyl quinate/shikimate 3′-hydroxylase (C3′H) and cinnamate 4-hydroxylase (C4H) genes in the lignin biosynthetic pathway of Eucalyptus urophylla x E grandis leads to improved sugar release. Biotechnol Biofuels 8:128. https://doi.org/10.1186/s13068-015-0316-x
Tamasloukht B, Wong Quai Lam MSJ, Martinez Y, Tozo K, Barbier O, Jourda C, Jauneau A, Borderies G, Balzergue S, Renou JP, Huguet S, Martinant JP, Tatout C, Lapierre C, Barrire Y, Goffner D, Pichon M (2011) Characterization of a cinnamoyl-CoA reductase 1 (CCR1) mutant in maize: effects on lignification, fibre development, and global gene expression. J Exp Bot 62:3837–3848. https://doi.org/10.1093/jxb/err077
Townsley BT, Sinha NR, Kang J (2013) KNOX1 genes regulate lignin deposition and composition in monocots and dicot. Front Plant Sci 4:121. https://doi.org/10.3389/fpls.2013.00121
Valdivia M, Galan JL, Laffarga J, Ramos JL (2016) Biofuels 2020: Biorefineries based on lignocellulosic materials. Microb Biotechnol 9:585–594. https://doi.org/10.1111/1751-7915.12387
Van Acker R, Vanholme R, Storme V, Mortimer JC, Dupree P, Boerjan W (2013) Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol Biofuels 6:46. https://doi.org/10.1186/1754-6834-6-46
Vanholme R, Storme V, Vanholme B, Sundin L, Christensen JH, Goeminne G, Halpin C, Rohde A, Morreel K, Boerjana W (2012) A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 24:3506–3529. https://doi.org/10.1105/tpc.112.102574
Vanholme R, De Meester B, Ralph J, Boerjan W (2019) Lignin biosynthesis and its integration into metabolism. Curr Opin Biotechnol 56:230–239. https://doi.org/10.1016/j.copbio.2019.02.018
Velez-Bermúdez IC, Salazar-Henao JE, Fornalé S, Löpez-Vidriero I, Franco-Zorrilla JM, Grotewold E, Gray J, Solano R, Schmidt W, Pagés M, Riera M, Caparros-Ruiz D (2015) A MYB/ZML complex regulates wound-induced lignin genes in Maize. Plant Cell 27:3245–3259. https://doi.org/10.1105/tpc.15.00545
Vicentini R, Bottcher A, Dos Santos BM, Dos Santos AB, Creste S, De Andrade Landell MG, Cesarino I, Mazzafera P (2015) Large-scale transcriptome analysis of two sugarcane genotypes contrasting for lignin content. PLoS ONE 10:e0134909. https://doi.org/10.1371/journal.pone.0134909
Voelker SL, Lachenbruch B, Meinzer FC, Jourdes M, Ki C, Patten AM, Davin LB, Lewis NG, Tuskan GA, Gunter L, Decker SR, Selig MJ, Sykes R, Himmel ME, Kitin P, Shevchenko O, Strauss SH (2010) Antisense down-regulation of 4CL expression alters lignification, tree growth, and saccharification potential of field-grown poplar. Plant Physiol 154:874–886. https://doi.org/10.1104/pp.110.159269
Wang H, Avci U, Nakashima J, Hahn MG, Chen F, Dixon RA (2010) Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc Natl Acad Sci U S A 107:22338–22343. https://doi.org/10.1073/pnas.1016436107
Wang JP, Chuang L, Loziuk PL, Chen H, Lin YC, Shi R, Qu GZ, Muddiman DC, Sederoff RR, Chiang VL (2015) Phosphorylation is an on/off switch for 5-hydroxyconiferaldehyde O-methyltransferase activity in poplar monolignol biosynthesis. Proc Natl Acad Sci U S A 112:8481–8486. https://doi.org/10.1073/pnas.1510473112
Wu Z, Wang N, Hisano H, Cao Y, Wu F, Liu W, Bao Y, Wang ZY, Fu C (2019) Simultaneous regulation of F5H in COMT-RNAi transgenic switchgrass alters effects of COMT suppression on syringyl lignin biosynthesis. Plant Biotechnol J 17:836–845. https://doi.org/10.1111/pbi.13019
Wuddineh WA, Mazarei M, Zhang J-Y, Turner GB, Sykes RW, Decker SR, Davis MF, Udvardi MK, Stewart CN (2016) Identification and overexpression of a knotted1-like transcription factor in switchgrass (Panicum virgatum L.) for lignocellulosic feedstock improvement. Front Plant Sci 7:520. https://doi.org/10.3389/fpls.2016.00520
Xiong W, Wu Z, Liu Y, Li Y, Su K, Bai Z, Guo S, Hu Z, Zhang Z, Bao Y, Sun J, Yang G, Fu C (2019) Mutation of 4-coumarate: Coenzyme A ligase 1 gene affects lignin biosynthesis and increases the cell wall digestibility in maize brown midrib5 mutants. Biotechnol Biofuels 12:82. https://doi.org/10.1186/s13068-019-1421-z
Xu C, Shen Y, He F, Fu X, Yu H, Lu W, Li Y, Li C, Fan D, Wang HC, Luo K (2019) Auxin-mediated Aux/IAA-ARF-HB signaling cascade regulates secondary xylem development in Populus. New Phytol 222:752–767. https://doi.org/10.1111/nph.15658
Yan L, Xu C, Kang Y, Gu T, Wang D, Zhao S, Xia G (2013) The heterologous expression in Arabidopsis thaliana of sorghum transcription factor SbbHLH1 downregulates lignin synthesis. J Exp Bot 64:3021–3032. https://doi.org/10.1093/jxb/ert150
Yu S, Kim H, Yun DJ, Suh MC, Lee B (2019) Post-translational and transcriptional regulation of phenylpropanoid biosynthesis pathway by Kelch repeat F-box protein SAGL1. Plant Mol Biol 99:135–148. https://doi.org/10.1007/s11103-018-0808-8
Zeng Y, Zhao S, Yang S, Ding SY (2014) Lignin plays a negative role in the biochemical process for producing lignocellulosic biofuels. Curr Opin Biotechnol 27:38–45. https://doi.org/10.1016/j.copbio.2013.09.008
Zhang X, Gou M, Liu CJ (2013) Arabidopsis kelch repeat F-Box proteins regulate phenylpropanoid biosynthesis via controlling the turnover of phenylalanine ammonia-lyase. Plant Cell 25:4994–5010. https://doi.org/10.1105/tpc.113.119644
Zhang J, Ge H, Zang C, Li X, Grierson D, Chen KS, Yin XR (2016) EjODO1, a MYB transcription factor, regulating lignin biosynthesis in developing loquat (Eriobotrya japonica) fruit. Front Plant Sci 7:1360. https://doi.org/10.3389/fpls.2016.01360
Zhang X, Gonzalez-Carranza ZH, Zhang S, Miao Y, Liu C, Roberts JA (2019) F-Box proteins in plants. Annu Plant Rev 2:1–21. https://doi.org/10.1002/9781119312994.apr0701
Zhong R, Richardson EA, Ye ZH (2007) The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19:2776–2792. https://doi.org/10.1105/tpc.107.053678
Acknowledgements
We gratefully acknowledge the support of Angela O’Keefe for planning the field trips, helping with harvest and keeping the lab running smoothly. We are also grateful to Jane Brownlee and Patrick Mason for their assistance with the field work. Special thanks to Fernando Correr for his advice on the differential expression analysis. We also thank the University of Queensland Research Computing Centre for their support in computing and data management.
Funding
This research was supported by The Australian Research Council via the ARC-Linkage funding scheme (Project Number LP160100939). Linkage partners include Sugar Research Australia and the Joint BioEnergy Institute (http://www.jbei.org) which is supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy. The funders had no role in experimental design, sample collection, data analysis or manuscript writing/editing.
Author information
Authors and Affiliations
Contributions
RH, FB, AF, and KHK conceived and designed the experiments. KHK and VP conducted the majority of the experiments with assistance from HC. KHK performed the data analysis and wrote the manuscript. RH, FB, BAS, JMG, AF, VP, HC and KHK critically revised the manuscript. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11103_2021_1136_MOESM1_ESM.docx
Supplementary file1 Additional file 1. Supplementary methods Statistical analyses. Table S1 Syringyl to guaiacyl (S/G) ratio of 22 sugarcane (Saccharum spp. hybrids) genotypes and their classification into high and low S/G ratio groups for the differential expression analysis. Letter groups were determined from a Tukey’s honestly significant difference test. Letter groups that differ indicate significant differences between genotypes for S/G ratio at a p-value <0.05. Table S2 RNA-Seq mapping statistics for high and low syringyl to guaiacyl (S/G) ratio sugarcane (Saccharum spp. hybrids) genotypes mapped against the SUGarcane Isoform-sequencing Transcriptome reference. Fig. S1 Mean expression of lignin biosynthesis transcripts differentially expressed between sugarcane (Saccharum spp. hybrids) genotypes with contrasting syringyl/guaiacyl (S/G) ratios (FDR p-value<0.01). Table S3 F-box protein transcripts tentatively involved in post-translational modification differentially expressed between sugarcane (Saccharum spp. hybrids) genotypes with contrasting syringyl/guaiacyl (S/G) ratios mapped against the SUGarcane Isoform-sequencing Transcriptome (SUGIT) reference (DOCX 39 kb)
11103_2021_1136_MOESM2_ESM.xlsx
Supplementary file2 Additional file 2. Complete list of significantly differentially expressed transcripts (FDR p-value ≤ 0.01) between sugarcane (Saccharum spp. hybrids) genotypes with contrasting syringyl to guaiacyl (S/G) ratios mapped against the SUGarcane Isoform-sequencing Transcriptome reference (XLSX 136 kb)
11103_2021_1136_MOESM3_ESM.docx
Supplementary file3 Additional file 3. FASTA-formatted sequences of significantly differentially expressed transcripts (FDR p-value ≤ 0.01) between sugarcane (Saccharum spp. hybrids) genotypes with contrasting syringyl to guaiacyl (S/G) ratios mapped against the SUGarcane Isoform-sequencing Transcriptome reference (DOCX 2039 kb)
11103_2021_1136_MOESM4_ESM.xlsx
Supplementary file4 Additional file 4. Complete list of single nucleotide polymorphisms associated with syringyl to guaiacyl (S/G) ratio in sugarcane (Saccharum spp. hybrids) with Gene Ontology functional annotations and NCBI BLAST descriptions for the SNP-containing transcripts (XLSX 238 kb)
Rights and permissions
About this article
Cite this article
Hodgson-Kratky, K., Perlo, V., Furtado, A. et al. Association of gene expression with syringyl to guaiacyl ratio in sugarcane lignin. Plant Mol Biol 106, 173–192 (2021). https://doi.org/10.1007/s11103-021-01136-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-021-01136-w