Abstract
Key message
The full-length transcriptome of alfalfa was analyzed with PacBio single-molecule long-read sequencing technology. The transcriptome data provided full-length sequences and gene isoforms of transcripts in alfalfa, which will improve genome annotation and enhance our understanding of the gene structure of alfalfa.
Abstract
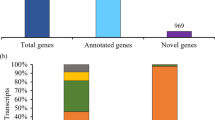
As an important forage, alfalfa (Medicago sativa L.) is world-wide planted. For its complexity of genome and unfinished whole genome sequencing, the sequences and complete structure of mRNA transcripts remain unclear in alfalfa. In this study, single-molecule long-read sequencing was applied to investigate the alfalfa transcriptome using the Pacific Biosciences platform, and a total of 113,321 transcripts were obtained from young, mature and senescent leaves. We identified 72,606 open reading frames including 46,616 full-length ORFs, 1670 transcription factors from 54 TF families and 44,040 simple sequence repeats from 30,797 sequences. A total of 7568 alternative splicing events was identified and the majority of alternative splicing events in alfalfa was intron retention. In addition, we identified 17,740 long non-coding RNAs. Our results show the feasibility of deep sequencing full-length RNA from alfalfa transcriptome on a single-molecule level.









Similar content being viewed by others
Data availability
We deposited the raw bam files of SMRT data in the Sequence Read Archives (SRA) of the National Center for Biotechnology Information (NCBI) under accession number SUB4116865. The Illumina RNA-Seq data was deposited in SRA under accession number SUB4113911.
Abbreviations
- ORF :
-
Open reading frame
- lncRNA :
-
Long non-coding RNA
- SSR :
-
Simple sequence repeat
- TF :
-
Transcript factor
- NGST:
-
Next-generation high-throughput sequencing technology
- SMRT :
-
Single molecule long reads sequencing technology
- AS:
-
Alterative splice
References
Abdel-Ghany SE, Hamilton M, Jacobi JL, Ngam P, Devitt N, Schilkey F, Ben-Hur A, Reddy AS (2016) A survey of the sorghum transcriptome using single-molecule long reads. Nat Commun 7:11706
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402
Bairoch A, Boeckmann B (1991) The SWISS-PROT protein sequence data bank. Nucleic Acids Res 19(Suppl):2247–2249
Barnes D (1980) Alfalfa. Hybrid Crop Plants. https://doi.org/10.2135/1980.hybridizationofcrops.c9
Chen SY, Deng FL, Jia XB, Li C, Lai SJ (2017) A transcriptome atlas of rabbit revealed by PacBio single-molecule long-read sequencing. Sci Rep 7:7648
Dilernia DA, Chien JT, Monaco DC, Brown MP, Ende Z, Deymier MJ, Yue L, Paxinos EE, Allen S, Tirado-Ramos A, Hunter E (2015) Multiplexed highly-accurate DNA sequencing of closely-related HIV-1 variants using continuous long reads from single molecule, real-time sequencing. Nucleic Acids Res 43(20):e129
Dowhan DH, Hong EP, Auboeuf D, Dennis AP, Wilson MM, Berget SM, O’Malley BW (2005). Steroid hormone receptor coactivation and alternative RNA splicing by U2AF(65)-related proteins CAPER alpha and CAPER beta. Mol Cell 17(3): 429–439
Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14(9):755–763
Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M (2014) Pfam: the protein families database. Nucleic Acids Res 42(D1):D222–D230
Foissac S, Sammeth M (2007) ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res 35(Web Server issue):W297–W299
Fu C, Hernandez T, Zhou C, Wang ZY (2015) Alfalfa (Medicago sativa L.). Methods Mol Biol 1223:213–221
Gordon SP, Tseng E, Salamov A, Zhang J, Meng X, Zhao Z, Kang D, Underwood J, Grigoriev IV, Figueroa M, Schilling JS, Chen F, Wang Z (2015) Widespread polycistronic transcripts in fungi revealed by single-molecule mRNA sequencing. PLoS ONE 10(7):e0132628
Guo AY, Chen X, Gao G, Zhang H, Zhu QH, Liu XC, Zhong YF, Gu X, He K, Luo J (2008) PlantTFDB: a comprehensive plant transcription factor database. Nucleic Acids Res 36(Database issue):D966–D969
Hackl T, Hedrich R, Schultz J, Förster F (2014) proovread: large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics 30(21):3004–3011
Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, Rattei T, Mende DR, Sunagawa S, Kuhn M, Jensen LJ, von Mering C, Bork P (2016) eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res 44(D1):D286–D293
Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32(Database issue):D277–D280
Kong L, Zhang Y, Ye Z-Q, Liu X-Q, Zhao S-Q, Wei L, Gao G (2007) CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35(Web Server issue):W345–W349
Koonin EV, Fedorova ND, Jackson JD, Jacobs AR, Krylov DM, Makarova KS, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Rogozin IB, Smirnov S, Sorokin AV, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA (2004) A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol 5(2):R7–R7
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22(13):1658–1659
Li Y, Dai C, Hu C, Liu Z, Kang C (2017) Global identification of alternative splicing via comparative analysis of SMRT- and Illumina-based RNA-seq in strawberry. Plant J 90(1):164–176
Liang M, Raley C, Zheng X, Kutty G, Gogineni E, Sherman BT, Sun Q, Chen X, Skelly T, Jones K, Stephens R, Zhou B, Lau W, Johnson C, Imamichi T, Jiang M, Dewar R, Lempicki RA, Tran B, Kovacs JA, Huang DW (2016) Distinguishing highly similar gene isoforms with a clustering-based bioinformatics analysis of PacBio single-molecule long reads. BioData Min 9:13
Liu W, Zhang Z, Chen S, Ma L, Wang H, Dong R, Wang Y, Liu Z (2016) Global transcriptome profiling analysis reveals insight into saliva-responsive genes in alfalfa. Plant Cell Rep 35(3):561–571
Liu W, Xiong C, Yan L, Zhang Z, Ma L, Wang Y, Liu Y, Liu Z (2017a) Transcriptome analyses reveal candidate genes potentially involved in al stress response in alfalfa. Front Plant Sci 8:26
Liu X, Mei W, Soltis PS, Soltis DE, Barbazuk WB (2017b) Detecting alternatively spliced transcript isoforms from single-molecule long-read sequences without a reference genome. Mol Ecol Resour 17(6):1243–1256
Marquez Y, Brown JW, Simpson C, Barta A, Kalyna M (2012) Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res 22(6):1184–1195
Mayjonade B, Gouzy J, Donnadieu C, Pouilly N, Marande W, Callot C, Langlade N, Munos S (2017) Extraction of high-molecular-weight genomic DNA for long-read sequencing of single molecules. Biotechniques 62(1)
Michael TP (2011) Exploring the Arabidopsis genome with long. single molecule PacBio reads. In Vitro Cell Dev Biol-Anim 47:S14–S14
Minoche AE, Dohm JC, Schneider J, Holtgrawe D, Viehover P, Montfort M, Sorensen TR, Weisshaar B, Himmelbauer H (2015) Exploiting single-molecule transcript sequencing for eukaryotic gene prediction. Genome Biol 16
Ning G, Cheng X, Luo P, Liang F, Wang Z, Yu G, Li X, Wang D, Bao M (2017) Hybrid sequencing and map finding (HySeMaFi): optional strategies for extensively deciphering gene splicing and expression in organisms without reference genome. Sci Rep 7:43793
Palusa SG, Ali GS, Reddy ASN (2007) Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J 49(6):1091–1107
Peng Z, Hu Y, Xie J, Potnis N, Akhunova A, Jones J, Liu Z, White FF, Liu S (2016) Long read and single molecule DNA sequencing simplifies genome assembly and TAL effector gene analysis of Xanthomonas translucens. BMC Genom 17:21
Postnikova OA, Hult M, Shao J, Skantar A, Nemchinov LG (2015) Transcriptome analysis of resistant and susceptible alfalfa cultivars infected with root-knot nematode Meloidogyne incognita. PLoS ONE 10(3):e0123157
Pyo CW, Vierra-Green C, Pyon YS, Eng K, Hall R, Hon L, Ranade S, Geraghty D (2014) Complete resequencing of extended genomic regions using fosmid target capture and single molecule real-time (Smrt) long read sequencing technology. Hum Immunol 75:5–5
Rashmi R, Manisha Sarkar V (1997) Cultivation of alfalfa (Medicago sativa L)". Anc Sci Life 17(2):117–119
Reddy AS (2007) Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu Rev Plant Biol 58:267–294
Rodet F, Lelong C, Dubos MP, Favrel P (2008) Alternative splicing of a single precursor mRNA generates two subtypes of Gonadotropin-releasing Hormone receptor orthologues and their variants in the bivalve mollusc Crassostrea gigas. Gene 414(1–2):1–9
Sharon D, Tilgner H, Grubert F, Snyder M (2013) A single-molecule long-read survey of the human transcriptome. Nat Biotechnol 31(11):1009
Song F, Li J, Fan X, Zhang Q, Chang W, Yang F, Geng G (2016a) Transcriptome analysis of Glomus mosseae/Medicago sativa mycorrhiza on atrazine stress. Sci Rep 6:20245
Song L, Jiang L, Chen Y, Shu Y, Bai Y, Guo C (2016b) Deep-sequencing transcriptome analysis of field-grown Medicago sativa L. crown buds acclimated to freezing stress. Funct Integr Genom 16(5):495–511
Steijger T, Abril JF, Engstrom PG, Kokocinski F, Consortium R, Hubbard TJ, Guigo R, Harrow J, Bertone P (2013) Assessment of transcript reconstruction methods for RNA-seq. Nat Methods 10(12):1177–1184
Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, Liu Y, Chen R, Zhao Y (2013) Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res 41(17):e166–e166
Tatusov RL, Galperin MY, Natale DA, Koonin EV (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28(1):33–36
The Gene Ontology, Ashburner CM, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. Nat Genet 25(1):25–29
Tilgner H, Raha D, Habegger L, Mohiuddin M, Gerstein M, Snyder M (2013) Accurate identification and analysis of human mRNA isoforms using deep long read sequencing. G3-Genes Genomes Genet 3(3):387–397
Tombacz D, Moldovan N, Balazs Z, Csabai Z, Snyder M, Boldogkoi Z (2017a) Genetic adaptation of porcine circovirus type 1 to cultured porcine kidney cells revealed by single-molecule long-read sequencing technology. Genome Announc 5(5):e01539–16
Tombacz D, Balazs Z, Csabai Z, Moldovan N, Szucs A, Sharon D, Snyder M, Boldogkoi Z (2017b) Characterization of the dynamic transcriptome of a herpesvirus with long-read single molecule real-time sequencing. Sci Rep 7:43751
Vembar SS, Seetin M, Lambert C, Nattestad M, Schatz MC, Baybayan P, Scherf A, Smith ML (2016) Complete telomere-to-telomere de novo assembly of the Plasmodium falciparum genome through long-read (> 11 kb), single molecule, real-time sequencing. DNA Res 23(4):339–351
Wang L, Park HJ, Dasari S, Wang S, Kocher J-P, Li W (2013) CPAT: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res 41(6):e74–e74
Wang B, Tseng E, Regulski M, Clark TA, Hon T, Jiao Y, Lu Z, Olson A, Stein JC, Ware D (2016a) Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat Commun 7:11708
Wang D, Khurshid M, Sun ZM, Tang YX, Zhou ML, Wu YM (2016b) Genetic engineering of alfalfa (Medicago sativa L.). Protein Pept Lett 23(5):495–502
Wang J, Zhao Y, Ray I, Song M (2016c) Transcriptome responses in alfalfa associated with tolerance to intensive animal grazing. Sci Rep 6:19438
Wang T, Wang H, Cai D, Gao Y, Zhang H, Wang Y, Lin C, Ma L, Gu L (2017) Comprehensive profiling of rhizome-associated alternative splicing and alternative polyadenylation in moso bamboo (Phyllostachys edulis). Plant J 91(4):684–699
Workman RE, Myrka AM, Wong GW, Tseng E, Welch KC, Timp W (2018) Single-molecule, full-length transcript sequencing provides insight into the extreme metabolism of the ruby-throated hummingbird Archilochus colubris. Gigascience 7(3):giy009
Wu TD, Watanabe CK (2005) GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21(9):1859–1875
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39(Web Server issue):W316–W322
Xu ZC, Peters RJ, Weirather J, Luo HM, Liao BS, Zhang X, Zhu YJ, Ji AJ, Zhang B, Hu SN, Au KF, Song JY, Chen SL (2015) Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant J 82(6):951–961
Xu QS, Zhu JY, Zhao SQ, Hou Y, Li FD, Tai YL, Wan XC, Wei CL (2017) Transcriptome profiling using single-molecule direct RNA sequencing approach for in-depth understanding of genes in secondary metabolism pathways of Camellia sinensis. Front Plant Sci 8:1205
Zhang P, Deng H, Mao FM, Liu YS (2013) Alterations of alternative splicing patterns of ser/arg-rich (SR) genes in response to hormones and stresses treatments in different ecotypes of rice (Oryza sativa). J Integr Agric 12(5):737–748
Zhang S, Shi Y, Cheng N, Du H, Fan W, Wang C (2015) De novo characterization of fall dormant and nondormant alfalfa (Medicago sativa L.) leaf transcriptome and identification of candidate genes related to fall dormancy. PLoS ONE 10(3):e0122170
Zhu FY, Chen MX, Ye NH, Shi L, Ma KL, Yang JF, Cao YY, Zhang YJ, Yoshida T, Fernie AR, Fan GY, Wen B, Zhou R, Liu TY, Fan T, Gao B, Zhang D, Hao GF, Xiao S, Liu YG, Zhang JH (2017) Proteogenomic analysis reveals alternative splicing and translation as part of the abscisic acid response in Arabidopsis seedlings. Plant J 91(3):518–533
Zhu J, Wang X, Guo L, Xu Q, Zhao S, Li F, Yan X, Liu S, Wei C (2018) Characterization and alternative splicing profiles of lipoxygenase gene family in tea plant (Camellia sinensis). Plant Cell Physiol 59:1765–1781
Acknowledgements
The program was supported by the National Natural Science Foundation of China (Grant Nos. 31601989 and 31672477). We acknowledge Jingjing Sui, Huaigen Xin and Dandan Chen from Biomarker Corporation (Beijing, China) for the facilities and expertise of the PacBio platform for libraries construction and sequencing.
Author information
Authors and Affiliations
Contributions
YC and LH conceived and designed the research. YC, JY and TG conducted experiments. ZM and LX analyzed data. YC and LH wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Communicated by Liebao Han.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chao, Y., Yuan, J., Guo, T. et al. Analysis of transcripts and splice isoforms in Medicago sativa L. by single-molecule long-read sequencing. Plant Mol Biol 99, 219–235 (2019). https://doi.org/10.1007/s11103-018-0813-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-018-0813-y