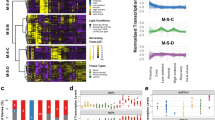
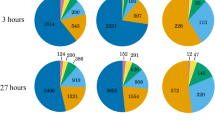
Abstract
During cold acclimation plants increase in freezing tolerance in response to low non-freezing temperatures. This is accompanied by many physiological, biochemical and molecular changes that have been extensively investigated. In addition, plants of many species, including Arabidopsis thaliana, become more freezing tolerant during exposure to mild, non-damaging sub-zero temperatures after cold acclimation. There is hardly any information available about the molecular basis of this adaptation. Here, we have used microarrays and a qRT-PCR primer platform covering 1,880 genes encoding transcription factors (TFs) to monitor changes in gene expression in the Arabidopsis accessions Columbia-0, Rschew and Tenela during the first 3 days of sub-zero acclimation at −3 °C. The results indicate that gene expression during sub-zero acclimation follows a tighly controlled time-course. Especially AP2/EREBP and WRKY TFs may be important regulators of sub-zero acclimation, although the CBF signal transduction pathway seems to be less important during sub-zero than during cold acclimation. Globally, we estimate that approximately 5 % of all Arabidopsis genes are regulated during sub-zero acclimation. Particularly photosynthesis-related genes are down-regulated and genes belonging to the functional classes of cell wall biosynthesis, hormone metabolism and RNA regulation of transcription are up-regulated. Collectively, these data provide the first global analysis of gene expression during sub-zero acclimation and allow the identification of candidate genes for forward and reverse genetic studies into the molecular mechanisms of sub-zero acclimation.







Similar content being viewed by others
References
Agarwal M, Hao Y, Kapoor A, Dong C-H, Fuji H, Zheng X, Zhu J-K (2006) A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 281:37636–37645
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300
Bieniawska Z, Espinoza C, Schlereth A, Sulpice R, Hincha DK, Hannah MA (2008) Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol 147:263–279
Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C et al (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43:956–963
Castonguay Y, Nadeau P, Laberge S (1993) Freezing tolerance and alteration of translatable mRNAs in alfalfa (Medicago sativa L.) hardened at subzero temperatures. Plant Cell Physiol 34:31–38
Chen L, Song Y, Li S, Zhang L, Zou C, Yu D (2012) The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta 1819:120–128
Chinnusamy V, Zhu J, Zhu J-K (2007) Cold stress regulation of gene expression in plants. Trends Plant Sci 12:444–451
Cui F, Brosch M, Sipari N, Tang S, Overmyer K (2013) Regulation of ABA dependent wound induced spreading of cell death by MYB108. New Phytol 200:634–640
Czechowski T, Bari R, Stitt M, Scheible W-R, Udvardi M (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38:366–379
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17
Espevig T, DaCosta M, Hoffmann L, Aamlid TS, Tronsmo AM, Clark BB, Huang B (2011) Freezing tolerance and carbohydrate changes of two Agrostis species during cold acclimation. Crop Sci 51:1188–1197
Espinoza C, Degenkolbe T, Caldana C, Zuther E, Leisse A, Willmitzer L, Hincha DK, Hannah MA (2010) The interaction between diurnal and circadian regulation results in dynamic metabolic and transcriptional changes during cold acclimation in Arabidopsis. PLoS ONE 5:e14101
Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14:1675–1690
Guy CL, Kaplan F, Kopka J, Selbig J, Hincha DK (2008) Metabolomics of temperature stress. Physiol Plant 132:220–235
Haake V, Cooke D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ (2002) Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 130:639–648
Hannah MA, Heyer AG, Hincha DK (2005) A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet 1:e26
Hannah MA, Wiese D, Freund S, Fiehn O, Heyer AG, Hincha DK (2006) Natural genetic variation of freezing tolerance in Arabidopsis. Plant Physiol 142:98–112
Herman EM, Rotter K, Premakumar R, Elwinger G, Bae R, Ehler-King L, Chen S, Livingston DP III (2006) Additional freeze hardiness in wheat acquired by exposure to -3 C is associated with extensive physiological, morphological, and molecular changes. J Exp Bot 57:3601–3618
Hincha DK, Espinoza C, Zuther E (2012) Transcriptomic and metabolomic approaches to the analysis of plant freezing tolerance and cold acclimation. In: Tuteja N, Gill SS, Toburcio AF, Tuteja R (eds) Improving Crop Resistance to Abiotic Stress. Wiley-Blackwell, Berlin, pp 255–287
Hong J-P, Takeshi Y, Kondou Y, Schachtman DP, Matsui M, Shin R (2013) Identification and characterization of transcription factors regulating Arabidopsis HAK5. Plant Cell Physiol 154:1478–1490
Hu Y, Jiang L, Wang F, Yu D (2013) Jasmonate regulates the INDUCER OF CBF EXPRESSION-C-REPEAT BINDING FACTOR/DRE BINDING FACTOR1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25:2907–2924
Hua J (2009) From freezing to scorching, transcriptional responses to temperature variation in plants. Curr Opin Plant Biol 12:568–573
Jaglo-Ottosen K, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF (1998) Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280:104–106
Kang HG, Kim J, Kim B, Jeong H, Choi SH, Kim EK, Lee HY, Lim PO (2011) Overexpression of FTL1/DDF1, an AP2 transcription factor, enhances tolerance to cold, drought, and heat stress in Arabidopsis thaliana. Plant Sci 180:634–641
Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotech 17:287–291
Kim YS, Park S, Gilmour SJ, Thomashow MF (2013) Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J 75:364–376
Knight H, Zarka DG, Okamoto H, Thomashow MF, Knight MR (2004) Abscisic acid induces CBF gene transcription and subsequent induction of cold-regulated genes via CRT promoter element. Plant Physiol 135:1710–1717
Kumar SV, Wigge PA (2010) H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140:136–147
Le MQ, Engelsberger WR, Hincha DK (2008) Natural genetic variation in acclimation capacity at sub-zero temperatures after cold acclimation at 4° C in different Arabidopsis thaliana accessions. Cryobiology 57:104–112
Licausi F, Ohme-Takagi M, Perata P (2013) APETALA2/Ethylene Response Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol 199:639–649
Liu ZQ, Yan L, Wu Z, Mei C, Lu K, Yu YT, Liang S, Zhang XF, Wang XF, Zhang DP (2012) Cooperation of three WRKY-domain transcription factors WRKY18, WRKY40, and WRKY60 in repressing two ABA-responsive genes ABI4 and ABI5 in Arabidopsis. J Exp Bot 63:6371–6392
Livingston DP III (1996) The second phase of cold hardening: freezing tolerance and fructan isomer changes in winter cereal crowns. Crop Sci 36:1568–1573
Livingston DP III, Van K, Premakumar R, Tallury SP, Herman EM (2007) Using Arabidopsis thaliana as a model to study subzero acclimation in small grains. Cryobiology 54:154–163
Lohse M, Nunes-Nesi A, Krüger P, Nagel A, Hannemann J, Giorgi FM, Childs L, Osorio S, Walther D, Selbig J et al (2010) Robin: an intuitive wizard application for R-based expression microarray quality assessment and analysis. Plant Physiol 153:642–651
Maruyama K, Takeda M, Kidokoro S, Yamada K, Sakuma Y, Urano S, Fujita M, Yoshiwara K, Matsukura S, Morishita Y et al (2009) Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol 150:1972–1980
Medina J, Catala R, Salinas J (2011) The CBFs: three Arabidopsis transcription factors to cold acclimate. Plant Sci 180:3–11
Monroy AF, Castonguay Y, Laberge S, Sarhan F, Vezina LP, Dhindsa RS (1993) A new cold-induced alfalfa gene is associated with enhanced hardening at sub-zero temperature. Plant Physiol 102:873–879
R Development Core Team (2010) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Redman JC, Haas BJ, Tanimoto G, Town CD (2004) Development and evaluation of an Arabidopsis whole genome Affymetrix probe array. Plant J 38:545–561
Rohde P, Hincha DK, Heyer AG (2004) Heterosis in the freezing tolerance of crosses between two Arabidopsis thaliana accessions (Columbia-0 and C24) that show differences in non-acclimated and acclimated freezing tolerance. Plant J 38:790–799
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M et al (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378
Sanchez DH (2013) Physiological and biotechnological implications of transcript-level variation under abiotic stress. Plant Biol 15:925–930
Schmid KJ, Törjek O, Meyer R, Schmuths H, Hoffmann MH, Altmann T (2006) Evidence for a large-scale population structure of Arabidopsis thaliana from genome-wide single nucleotide polymorphism markers. Theor Appl Genet 112:1104–1114
Shinozaki K, Yamaguchi-Shinozaki K, Seki M (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 6:410–417
Skinner DZ (2009) Post-acclimation transcriptome adjustment is a major factor in freezing tolerance of winter wheat. Funct Integr Genomics 9:513–523
Steponkus PL (1984) Role of the plasma membrane in freezing injury and cold acclimation. Annu Rev Plant Physiol 35:543–584
Thalhammer A, Hincha DK, Zuther E (2014) Measuring freezing tolerance: electrolyte leakage and chlorophyll fluorescence assays. In: Hincha DK, Zuther E (eds) Methods in molecular biology. Springer, New York, pp 15–24
Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50:571–599
Thomashow MF (2010) Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol 154:571–577
Törjek O, Berger D, Meyer RC, Müssig C, Schmid KJ, Rosleff Sörensen T, Weisshaar B, Mitchell-Olds T, Altmann T (2003) Establishment of a high-efficiency SNP-based framework marker set for Arabidopsis. Plant J 36:122–140
Usadel B, Nagel A, Steinhauser D, Gibon Y, Bläsing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F et al (2006) PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinf 7:535
van Buskirk HA, Thomashow MF (2006) Arabidopsis transcription factors regulating cold acclimation. Physiol Plant 126:72–80
van Leeuwen H, Kliebenstein DJ, West MAL, Kim K, van Poecke R, Katagiri F, Michelmore RW, Doerge RW, St. Clair DA (2007) Natural variation among Arabidopsis thaliana accessions for transcriptome response to exogenous salicylic acid. Plant Cell 19:2099–2110
Vogel JT, Zarka DG, van Buskirk HA, Fowler SG, Thomashow MF (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41:195–211
Weigel D (2012) Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiol 158:2–22
Xin Z, Browse J (2000) Cold comfort farm: the acclimation of plants to freezing temperatures. Plant Cell Environ 23:893–902
Zhen Y, Ungerer MC (2008a) Clinal variation in freezing tolerance among natural accessions of Arabidopsis thaliana. New Phytol 177:419–427
Zhen Y, Ungerer MC (2008b) Relaxed selection on the CBF/DREB1 regulatory genes and reduced freezing tolerance in the Southern range of Arabidopsis thaliana. Mol Biol Evol 25:2547–2555
Zhou MQ, Shen C, Wu LH, Tang KX, Lin J (2011a) CBF-dependent signaling pathways: a key responder to low temperature stress in plants. Crit Rev Biotechnol 31:186–192
Zhou X, Jiang Y, Yu D (2011b) WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol Cells 31:303–313
Zuther E, Schulz E, Childs LH, Hincha DK (2012) Natural variation in the non-acclimated and cold-acclimated freezing tolerance of Arabidopsis thaliana accessions. Plant Cell Environ 35:1860–1878
Acknowledgments
MQL was supported by a PhD fellowship from the Vietnamese Ministry of Education and Training and MP by a Postdoctoral fellowship from the Carlsberg Foundation (Denmark).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Mai Q. Le and Majken Pagter have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2014_256_MOESM1_ESM.tif
Score plots from PCA of the Ct values measured by qRT-PCR of transcripts from TF genes (left panel) and of the signal intensities from microarray hybridization experiments (right panel) with leaves of Arabidopsis thaliana accessions Columbia-0 (□), Rschew (○) and Tenela (Δ). Plants were cold acclimation at 4 °C for two weeks (black). Left panel: leaves sub-zero acclimated at -3 °C for 1 h (red), 2 h (blue), 3 h (green) or 8 h (yellow). Right panel: sub-zero acclimated for 8 h (red), 1 d (blue) or 3 d (green). Each symbol represents one replicate. Replicates which were excluded from further analysis are encircled (TIFF 1581 kb)
11103_2014_256_MOESM5_ESM.tif
A complete PageMan display of all significantly regulated bins and sub-bins during different durations of sub-zero acclimation of leaves of cold acclimated plants of the Arabidopsis thaliana accessions Col-0, Rsch and Te. Normalized gene expression values were subjected to an overrepresentation analysis to identify functional bins that contained significantly more or less regulated genes than expected by chance. Blue color indicates significant enrichment of up- or down-regulated genes, red indicates significant depletion (TIFF 296 kb)
11103_2014_256_MOESM9_ESM.tif
Time dependence of sub-zero acclimation at -3ºC in the Arabidopsis thaliana accessions Columbia-0, Rschew and Tenela. Plants were cold acclimated for two weeks at 4 ºC (CA). Detached leaves were then sub-zero acclimated at -3ºC for 1 d and 3 d (SZA). At the end of the acclimation period, leaves were frozen and thawed to determine the LT50 values. The bars denote mean ± SE from five biological replicates containing leaves from three plants each. The significance of the increase in freezing tolerance after sub-zero acclimation, compared to plants acclimated at 4 °C, was determined by t test and is indicated by asterisks (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001) (TIFF 102 kb)
Rights and permissions
About this article
Cite this article
Le, M.Q., Pagter, M. & Hincha, D.K. Global changes in gene expression, assayed by microarray hybridization and quantitative RT-PCR, during acclimation of three Arabidopsis thaliana accessions to sub-zero temperatures after cold acclimation. Plant Mol Biol 87, 1–15 (2015). https://doi.org/10.1007/s11103-014-0256-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-014-0256-z