Abstract
Promoter elements that contribute to high light (HL) induction of the Arabidopsis ELIP1 gene were defined using a transgenic promoter-reporter system. Two adjacent SORLIP1 elements (double SORLIP1, dSL) were found to be essential for HL induction of a GUS reporter gene. The dSL element was also found to be essential for HL induction conferred by the ELIP2 promoter. SORLIP1 elements were enriched in ELIP promoters throughout the plant kingdom, and showed a clade-specific pattern of gain or loss that suggested functionality. In addition, two G-box elements were found to redundantly contribute to HL induction conferred by the ELIP1 promoter.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Early light-induced proteins (ELIPs) were first identified as genes rapidly transcribed after etiolated seedlings were transferred from the dark to the light (Meyer and Kloppstech 1984). ELIPs are members of the light harvesting complex (LHC) superfamily and have three transmembrane domains that traverse the thylakoid membrane. They bind chlorophyll a and lutein (Adamska et al. 1999) and are widely distributed throughout the plant kingdom (Adamska 1997). ELIP genes are expressed at sparse levels under low light (LL) conditions, but the mRNA quickly becomes abundant in response to high light (HL). ELIP proteins can be detected in the thylakoid membrane within 2 h of HL exposure where they associate with the light harvesting complex of PSII (Heddad and Adamska 2002). ELIP proteins are then degraded soon after the return to LL (Adamska et al. 1993). The rapid responsiveness to HL is conserved throughout the plant kingdom (Heddad and Adamska 2002; Ensminger et al. 2004) and is distinct from the light responsiveness of other LHC family members (Klimmek et al. 2006).
Although sequence and expression patterns are highly conserved, a mechanism of action for ELIPs has not been elucidated by genetic analyses. Arabidopsis thaliana contains two ELIP genes (ELIP1, At3g22840 and ELIP2, At4g14690). Overexpression of each ELIP gene rescued the photosensitivity of the pleiotropic chaos mutant, which lacks cSRP43, and is defective in one of many thylakoid protein insertion pathways. The chaos mutant had higher levels of uncoupled chlorophylls, and the rescue by ELIPs suggested a function in sequestering unbound chlorophylls formed during HL (Hutin et al. 2003). However, double elip1elip2 null mutants displayed no changes in PSII photoinhibition, lipid peroxidation, or qE (nonphotochemical quenching) under HL conditions. The only differences were decreased accumulation of chlorophyll during greening and a decrease in zeaxanthin after HL treatment (Rossini et al. 2006). Overexpression of ELIP2 resulted in decreased chlorophyll accumulation due to a reduction in chlorophyll synthesis, predominantly at the Mg-chelation step (Tzvetkova-Chevolleau et al. 2007). Both knockout and overexpression of ELIPs resulted in decreased chlorophyll levels suggesting a complex relationship between ELIPs and chlorophyll synthesis/accumulation.
Besides HL responsiveness, a Genevestigator perturbations analysis (Hruz et al. 2008) showed significant increases in ELIP1 and ELIP2 expression in response to abiotic stresses such as UV-B (Genevestigator ID# AT-00528), cold (AT-00467), heat (AT-00179), drought (AT-00292), hypoxia (AT-00447), and anoxia (AT-00158). Additionally, ELIPs were induced in response to Pseudomonas syringae systemic infection, a biotic stress (AT-00363). Red (AT-00492), far-red (FR, AT-00109), and blue (AT-00109) light also stimulated ELIP expression. Interestingly, the response to red light still occurred in the phyABCDE mutant (AT-00601) suggesting the red light response is independent of phytochrome. Despite the inconclusive genetic results described above, these conserved, rapidly activated genes must play an important role in response to light as well as abiotic and biotic stresses.
Photoinhibition within the chloroplast correlates to ELIP gene transcription in the nucleus (Heddad et al. 2006), thus the rapid HL-induced expression of ELIP genes suggests the operation of chloroplast-nuclear retrograde signaling. Numerous pathways for retrograde signaling have been identified (Kleine and Leister 2013; Kleine et al. 2009), but none explain the rapid induction of ELIP genes. Reactive oxygen species are formed under HL, but ELIPs are not induced by superoxide or H2O2 (Gadjev et al. 2006; op den Camp et al. 2003; Van Aken and Whelan 2012). The carotenoid biosynthesis inhibitor norflurazon activates ELIP gene expression, but this activation still occurs in gun1 and gun5, suggesting independence from the tetrapyrrole retrograde signaling pathway (Brusslan and Peterson 2002; Koussevitzky et al. 2007) and ABI4 (Leon et al. 2013). Heme has recently been shown to be a retrograde signaling molecule, and a small dampening of ELI3 (the Chlamydomonas ELIP gene) induction occurs upon bilin feeding in the green algae Chlamydomonas, however ELI3 induction after a dark to light transition is normal in heme oxygenase mutants that cannot synthesize bilin (Duanmu et al. 2013). Furthermore, ELIP expression does not change in distal leaves during systemic acquired acclimation (Rossel et al. 2007). ELIP2 mRNA levels were higher in sal1 mutants that cannot produce the retrograde signaling PAP phosphonucleotide, however fold induction in response to HL was normal (Estavillo et al. 2011). It thus appears that ELIPs may be induced by a novel retrograde pathway.
Towards understanding ELIP retrograde signaling, the pea ELIP promoter was studied to identify important cis elements. Two well-known light regulatory elements (G-box and GT1) located approximately 120 bp from the start of transcription were implicated in ELIP induction when etiolated seedlings were exposed to light. These regions were protected from DNAse digestion by nuclear extracts from both etiolated and light-treated seedlings (Blecken et al. 1994). Both Arabidopsis ELIP genes were found to be induced early in response to FR light (Tepperman et al. 2001), and were included in an enumerative screen for promoter elements enriched in early FR-responsive genes (Hudson and Quail 2003). Novel sequences over-represented in light-induced promoters (SORLIP) were identified, with the most highly enriched sequence being SORLIP1 (GCCAC).
In this study, ELIP promoter elements that conferred HL responsiveness to a reporter gene were identified using site-directed mutagenesis of full-length promoter-reporter constructs stably integrated into the Arabidopsis genome. Two G-boxes in the ELIP1 promoter were found to redundantly contribute to HL responsiveness. In addition, a cis-region containing double SORLIP1 elements (dSL) was shown to be required for HL responsiveness for both ELIP1 and ELIP2 promoters.
Materials and methods
Plant material and growth conditions
The Arabidopsis thaliana (L.) Heynh. (ecotype Columbia or Landsberg erecta) plants used in this study were grown in a Percival growth chambers on Sunshine Mix #1 soil (Sun Gro Horticulture Distribution Inc.) under controlled conditions of light, namely LL (≈60 μmol photons m−2 s−1, 20 h light: 4 h dark) and temperature (23 °C). Light stress was imposed by transferring 21 day-old plants (n > 25 T2 seedlings) for 4 h to a Percival E356HO growth chamber (HL ≈ 900 μmol photons m−2 s−1). HL treatments were performed at the end of the 4 h dark period.
ELIP constructs and Agrobacterium-mediated plant transformation
The ELIP1 Wild Type (WT) promoter fragment (1,081 bp = −984 bp to +97 bp of the 5′-UTR) and the ELIP2 WT promoter fragment (954 bp = −883 bp to +71 bp of the 5′-UTR) were amplified by PCR from A. thaliana (ecotype Columbia) genomic DNA (Fig. 1). The primers used for ELIP1 were: ELIP1-forward: 5′-GGAATTCGAAACGACCGTAAATATTACC-3′ and ELIP1-reverse: 5′-GCGGATCCCTAGTG TGAGAGAAATTAAG-3′, and for ELIP2: ELIP2-SalI-Fw: 5′-GGGTCGACACAGCGCACGTAGGAGAATT-3′ and ELIP2-Rev: 5′-GCGGATCCAAGCCGAAGATCGATGAAGAAG-3′, introducing an EcoRI and BamHI and a SalI and BamHI restriction sites, respectively. These restriction sites were used to clone the promoter fragment into pBluescript II KS plasmid (pKS) (Stratagene, Inc.).
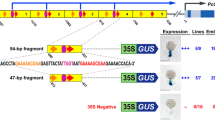
ELIP1 and ELIP2 promoter sequences. Sequence characteristics of a ELIP1- At3g22840 promoter (from −984 to +97 bp) and b ELIP2-At4g14690 promoter (from −883 to +71 bp). Upper case letters correspond to the promoter region and lower case letters correspond to the 5′-UTR. The start of translation (ATG) is underlined and in bold. Below the cis-regions that have been subjected to site-directed mutagenesis in this study (in bold and underlined) are their names and position relative to the start of transcription
After nucleotide sequence analysis to confirm the integrity of the WT promoter sequence, point mutations were introduced in potential cis-acting regions by using the QuickChange® Site Direct Mutagenesis Kit (Stratagene, Inc.). The list of primers used for the site-directed mutagenesis is found in Supplemental Table 1. In the case of the dSL mutated promoter, the first two point mutations in the dSL region were obtained using the primers SL1 (ELIP1) or SL1 (ELIP2). After sequencing and confirming the mutated sequence, the last two point mutations were introduced by using the primers dSL (ELIP1) or dSL (ELIP2) as appropriate. The SL2 (ELIP1) primers were used to create mutations in the second SORLIP1 element, by itself.
All mutated promoter constructs were verified by sequencing, and subcloned into the binary vector pBI101.1 (Clontech Laboratories Inc.) upstream of the GUS gene (uidA) by using the restriction sites SalI and BamHI. After confirming the integrity of the binary vectors containing the promoter fragment by restriction analysis, these were introduced into Agrobacterium tumefaciens (strain GV3101) by electroporation.
Wild type Arabidopsis plants were transformed using the floral dip transformation method (Clough and Bent 1998). T1 seeds were collected and then germinated in vitro on medium containing Linsmaier and Skoog salts (Caisson Laboratories Inc.), 20 g L−1 sucrose, 7 g L−1 agar (Phytablend™, Caisson Laboratories Inc.), 50 mg L−1 kanamycin and 50 mg L−1 carbenicillin. Antibiotic-resistant seedlings were transferred to soil and T2 seeds were collected from individual lines and used directly for experimentation.
The T2 plants used in the GUS Assay were also analyzed by PCR and sequencing. Genomic DNA was isolated using DNAzol according to manufacturer’s instructions except that volumes were decreased by 50 % (Invitrogen Inc.). In order to amplify part of the pBI101.1 plasmid and the entire full-length promoter region, the following primers were used: pBI-GUS reverse: 5′-ATGCCCACAGGCCGTCG-3′ and the PBI101.1 forward primer previously listed. This region was then sequenced to confirm that each line had the expected mutations. A partial segment of the GUS gene was also amplified in DNA extracted from T2 plants using the following primers: GUS-mid forward: 5′-AAGCCAGACAGAGTG TGATATC-3′ and the GUS-mid reverse: 5′-ATCAATCACCACGATGCCATG-3′. This region was amplified to be certain no deletions of the GUS gene had occurred during the transformation procedure.
Quantitative fluorometric GUS Assay
T2 seeds from each individual line were sown onto moist soil in 2 separate pots (n > 25 seeds per pot) and stratified at 4 °C for 3 days. The pots were then transferred to the growth chamber under the controlled conditions described above. After the LL and HL treatments, three separate samples of 100 mg of leaves were harvested per pot and treatment, containing different T2 plants of the same line. Two samples were used to perform the fluorometric Gus assay according to (Jefferson et al. 1987) and the remaining sample was used for RNA extraction. Briefly, leaves were directly ground in 500 μL of extraction buffer. After centrifugation, 50 μl of the supernatant was added to 500 μl of the assay buffer containing the substrate 4-methylumbelliferyl-β-D-glucuronide (MUG) (HACH Inc.) and incubated at 37 °C for 30 min. After 30 min, the reaction was stopped with 900 μl of stop buffer (0.2 M Na2CO3) and fluorescence due to the product 4-methyl-umbelliferone (4-MU) of the β-glucuronidase activity was measured with the DyNA Quant™ 200 fluorometer (Hoefer Pharmacia Biotech, Inc.). Before measurements, the fluorometer was calibrated with freshly prepared 1 μM 4-MU (Sigma-Aldrich, Inc.) standard and set to 500 relative fluorescence units (RFU). Protein concentration of plant extracts was determined by the Bio-Rad Protein Assay (Bio-Rad, Inc.), RFU values were normalized to the protein concentration in individual samples and Gus Activity was expressed as nmoles min−1 mg−1. Data were expressed as HL/LL fold induction.
Real time qPCR
RNA was isolated using Trizol reagent (Invitrogen Inc.), and cDNA was synthesized using random hexamers (Operon Biotechnologies Inc.) and MMLV reverse transcriptase (New England Biolabs Inc.). cDNA was diluted 1:3 prior to real-time PCR. Real time PCR amplification was performed in an MX3000P real-time PCR machine (Stratagene, Inc.) using 2× SYBR Green mix (AB Gene Inc.) in a total volume of 12 μL. PCR reactions all used a 61 °C annealing temperature, and dissociation curves were done to check for primer-dimers. The primers were as follows: ACT2-F: 5′-GGCGACTTGACAGAGAAGAA; ACT2-R: 5′-TGGAAAGAAAGAGCGGAAGA; Gus-Fw1: GAACTGAACTGGCAGACTATCCC; Gus-Rev1: 5′-TCGGCGTGGTGTAGAGCATTAC; Elip1-qRT-Fw: 5′-AAGGTGGGACACTCGTCTAAG; Elip1-qRT-Rv: 5′-GTGTTTTTAACCCGAAGTTTC; Elip2-qRT-Fw2: 5′-CCACAAATGCCACAGTCTC, Elip2-qRT-Rv2: 5′-CTCCAAACTTCGTACTCACC.
Statistical analysis
The non-parametric Mann–Whitney test (Mann and Whitney 1947) was used to determine significant differences in pairwise comparisons for GUS activity and mRNA analyses.
Results
Identification of ELIP promoter elements conferring HL induction
To define promoter elements that play a role in HL-induced ELIP1 expression, site-directed mutagenesis was performed on the ELIP1 promoter (ELIP1p) using a region that extended 984 bp upstream from the start of transcription and included 97 bp of the 115 bp 5′-UTR (Fig. 1). This region was cloned upstream of the GUS reporter gene and conferred a strong induction of GUS activity after 4 h of HL treatment (ELIP1 WT, Fig. 2). ELIP1 mRNA showed maximal induction after 3 h of HL exposure, and a 4 h HL exposure was utilized in all experiments to permit accumulation of GUS protein (Supplemental Figure 1). Numerous well-defined light regulatory elements (LREs) (Arguello-Astorga and Herera-Estrella 1998; Higo et al. 1999; Kuhlemeier et al. 1987) are located in the ELIP1 promoter: CAAT at -122 (relative to the start of transcription), GT1-like at -142, G-box at -175, GATA at -205 and upstream G-box (UpG-box) at -554. Full-length promoter regions with the LRE-element nucleotide changes shown in Table 1 were constructed as single, double and triple mutants, introduced into Arabidopsis via Agrobacterium floral dip transformation, and individual transgenic lines were selected and tested for HL induction of GUS activity in the T2 generation. Figure 2 shows the HL/LL fold induction for each construct (n = 20–25 transgenic lines). Most LREs did not significantly affect GUS HL/LL induction, however double mutations in both the G-box and UpG-box significantly (p = 0.0010) decreased GUS HL/LL induction in comparison to ELIP1 WT, suggesting the additive importance of these two elements. A small and slightly significant difference (p = 0.0243) was observed upon mutation of both the G-box and GATA elements, however a triple mutant (G-box, GATA, and CAAT) was not significantly different than ELIP1 WT. These findings suggest the GATA box has a small positive effect, while the CAAT box has a small negative effect on ELIP1 promoter activity.
GUS activity in ELIP1p-GUS transgenic lines. A 984 bp region of ELIP1p was fused to the GUS reporter gene (ELIP1 WT). Site-directed mutants of the 984 bp region were generated and also fused to the GUS reporter gene. Transgenic lines were generated and tested for GUS activity in LL and after 4 h of HL to produce the HL/LL fold induction. 20–25 transgenic lines were analyzed for each construct and error bars indicate the interquartile range (IQR). The location and sequence of promoter elements are shown in Fig. 1 while the nucleotide changes for each element are shown in Table 1. The SL1 and SL2 mutants change one of the two SORLIP1 elements in the dSL region. Pairwise comparisons between ELIP1 WT and each mutant construct were evaluated for statistical significance using the Mann–Whitney test. *p value 0.0500–0.0100, **p value 0.009–0.001, ***p value 0.0009 or below, but greater than 0
In order to find additional elements, ELIP1p and ELIP2p were analyzed using the Arabidopsis Promoter Element Discovery Tools (http://stan.cropsci.uiuc.edu/tools.php), and the SORLIP1 element (GCCAC) was found to be over-represented (p = 1.80e−04). Interestingly, a near perfect duplication of the SORLIP1 element (AGGCCAC GCCAT) within a completely conserved 12 bp region was found at −676 of ELIP1 and −581 of ELIP2. This 12 bp region is only found in the −1,000 region of one other expressed Arabidopsis gene, At2g38530, which encodes a stress-induced lipid transfer protein. This element was named double SORLIP1 (dSL), and subjected to site-directed mutagenesis of either one or both SORLIP1 elements (Table 1). ELIP1p with site-directed mutations singly disrupting the dSL SORLIP1 elements (SL1 and SL2) did not display significantly reduced HL/LL induction of GUS activity, however ELIP1p with both dSL SORLIP1 elements disrupted did show a significant reduction in HL/LL induction of GUS activity (p < 0.00009, Fig. 2). dSL mutants in combination with one or both G-box elements did not further decrease HL/LL induction of GUS activity suggesting that G-boxes and dSL are not additive.
Mutations in promoter regions can lead to complete inactivation. To demonstrate this had not occurred for dSL mutants, LL GUS activity levels were plotted in relation to HL/LL GUS fold induction in Supplemental Figure 2. Although the highest LL activity levels were observed for a subset of ELIP1p WT transgenic lines, many lines with greater than 1 nmole min−1 mg−1 of GUS activity were observed in the ELIP1p dSL mutant transgenic lines.
For a subset of constructs, tissue grown for GUS activity assays was also harvested for RNA extraction, and native ELIP1 and GUS mRNA were quantified by real-time qPCR using ACT2 as a reference (Livak and Schmittgen 2001). Expression of the native ELIP1 served as an internal control for HL induction and any samples that had native ELIP1 induction levels less than twofold were removed from the statistical analysis. Figure 3 shows the HL/LL fold induction for ELIP1p-GUS mRNA, and significant differences compared to WT were observed for the G-box UpG-box double mutant (p = 0.0002) as well as each single SORLIP1(p = 0.0009 for SL1 and 0.0006 for SL2). The dSL mutant and the dSL mutant combined with G-box promoter mutants were highly significantly different than WT (p < 0.00009 in all cases). Overall, the mRNA data show that the dSL is required for HL induction and that each SORLIP1 element within the dSL contributes to HL/LL fold induction along with the G-box and the UpG-box.
GUS mRNA levels in ELIP1p-GUS transgenic lines. ELIP1p-GUS lines were tested for RNA expression by real-time qPCR harvested from the same tissue used for GUS activity measurements. Both GUS and native ELIP1 mRNA levels were quantified and samples that displayed a less than twofold induction of native ELIP1 were removed from the analysis. GUS mRNA levels with a significantly lower HL/LL fold induction than ELIP1p WT are shown in white bars. The number of transgenic lines analyzed varied among constructs: ELIP1 WT (15), G-box-UpG-box (14), SL1 (8), SL2 (20), dSL (26), dSL-G-box (19), dSL-G-box-UpG-box (16). Error bars represent IQR. Statistical analysis and p values are as indicated for Fig. 2
If the dSL element is important for HL induction, it would be expected to play a similar role in the HL-inducible ELIP2 promoter, where it is present in a similar position (Fig. 1). Site directed mutagenesis was carried out in the context of a full-length ELIP2p (−883 to +71 of an 80 bp 5′-UTR), and both ELIP2p WT and ELIP2p dSL mutant promoters were cloned adjacent to the GUS reporter. Transgenic lines, were treated with HL and ELIP2p-GUS and native ELIP2 mRNA were quantified and any samples that had native ELIP2 induction levels less than twofold were removed from the statistical analysis. A significant decrease in HL/LL fold induction of GUS mRNA for the ELIP2p dSL mutant compared to the ELIP2p WT promoter construct was observed (p < 0.00009). These data show that the dSL element is required for ELIP2p to confer HL/LL induction on a reporter construct (Fig. 4).
The dSL element of ELIP2 is required for HL induction. ELIP2 WT (n = 14) and dSL mutant (n = 15) transgenic lines were tested for native ELIP2 and GUS mRNA expression by real-time qPCR. Samples that displayed a less than twofold induction of native ELIP2 were removed from the analysis. Error bars represent IQR. Statistical analysis and p values are as indicated for Fig. 2
To view the variability of the transgenic lines, GUS mRNA induction was plotted against native ELIP1 mRNA levels in Supplemental Figure 3a. The bulk of ELIP1p dSL mutant lines were clustered towards the y-axis, while many ELIP1p WT lines showed high HL/LL induction of GUS mRNA. A similar analysis with the ELIP2p constructs is shown in Supplemental Figure 3b. The high variability is most likely due to random integration of different copies of T-DNA constructs into more active and less active chromatin regions (Butaye et al. 2005).
SORLIP1 elements in ELIP promoter regions throughout the plant kingdom
To determine the distribution of SORLIP1 elements in ELIP promoter regions, 57 ELIP gene promoters from non-vascular as well as vascular plants were scanned for GCCAC sequences located 1,500 bp upstream from the start of translation. Most ELIP promoters contained 1–3 SORLIP1 elements (Fig. 5), however only the Arabidopsis genes contained the dSL element. When the frequency of SORLIP1 elements was compared to the random occurrence of any 5 bp region, SORLIP1 elements were found to be 1.5 fold enriched. This number is likely an underestimate since promoter regions tend to be AT-rich (Morey et al. 2011). ELIP genes were placed into a phylogenetic tree using Mesquite which displays the presence/absence of a trait (Maddison and Maddison 2011). The trait tracked was the presence of one or more SORLIP1 elements (Fig. 6). ELIP promoters with SORLIP1 elements were widely distributed among moss, monocots and dicots. For species with many ELIP paralogs, such as Eucalyptus grandis, specific clades had lost SORLIP1 elements suggesting functional diversification and not just random gain/loss, however gain/loss was more widely distributed for the Physcomitrella patens ELIP paralogs.
SORLIP1 elements in ELIP promoters throughout the plant kingdom. 37 ELIP promoters throughout the plant kingdom were identified and 1,500 bp upstream from the start of translation was scanned for SORLIP1 elements (GCCAC). The number of promoters (y-axis) with different numbers of SORLIP1 elements (x-axis) is shown. SORLIP1 elements were enriched 1.5 fold in ELIP promoter regions compared to random occurrence
SORLIP1 element gain/loss in ELIP promoter regions throughout the plant kingdom. ELIP promoters were scanned for SORLIP1 elements (GCCAC) and a phylogenetic tree was generated using Mesquite. Black signifies presence of at least one SORLIP1 element. The probability of a common ancestor having one or more SORLIP1 elements is shown by the proportion of black in common ancestor circles. The numbers represent maximum likelihood values supporting the ancestral proportions
Discussion
A transgenic promoter-reporter system was used to define elements within ELIP1p that are essential for responsiveness to HL. Defining these elements provides endpoint information on the retrograde signaling mechanism that regulates ELIP gene expression in response to HL. Large numbers of transgenic lines (14–27) were used to overcome the position effects that lead to high variability in reporter gene expression. Many classic LREs (GATA and CAAT) were found to only marginally contribute to HL activation by ELIP1p. However, two G-boxes, located at −175 and −554, redundantly increased HL/LL fold induction of GUS mRNA and activity. G-boxes have been implicated in HL activation previously (Blecken et al. 1994), however the extent of their contribution is unclear since microarray experiments show strong HL-induction of both ELIP1 and ELIP2 in the hy5 mutant, which encodes a bZIP transcription factor that binds to G-boxes (AT-00246) (Chattopadhyay et al. 1998). It is possible that other G-box binding TFs are involved in HL induction. The previous analysis of the pea ELIP promoter suggested that the GT1 element may be important (Blecken et al. 1994), but our study did not support a role for the ELIP1 GT1-like element. The pea study used different conditions to activate ELIP expression (etiolated seedlings transferred to low light as opposed to mature leaves exposed to HL) and utilized promoter deletions instead of site-directed mutagenesis of the full-length promoter.
GUS activity and mRNA levels had a significantly reduced response to HL when the dSL element was subjected to site-directed mutagenesis. The dSL element consists of two adjacent SORLIP1 elements, with the second one having a single nucleotide substitution, in a 12 bp region conserved between ELIP1p and ELIP2p. Mutagenesis of each SORLIP1 element resulted in lower HL induction of GUS mRNA, however a significant difference was not observed for GUS activity. Quantification of mRNA is a more direct measurement of promoter activity, and is more likely to reflect the importance of promoter elements. The ELIP1 dSL mutant promoters consistently showed a reduction in reporter induction after HL exposure, which was not reversed or strengthened by mutation in other elements. In addition, the dSL element was required for HL induction in ELIP2p. Taken together, our transgenic promoter-reporter analysis identified SORLIP1 elements to be essential for HL induction conferred on reporters for both Arabidopsis ELIP promoters. Interestingly, the minimal region that could confer light responsiveness (−228 to −74) in the earlier pea study did contain one SORLIP1 element at −110 (Kolanus et al. 1987), thus the pea study could also support a role of SORLIP1 elements in ELIP expression.
ELIP promoters throughout the plant kingdom were scanned for SORLIP1 and dSL elements. dSL elements were only found in the A. thaliana ELIP promoters, but SORLIP1 elements were distributed widely and a low level of enrichment (1.5×) was estimated based on the random occurrence of the five bp sequence. Two ELIP promoters had 10 SORLIP1 elements (Setaria Si031233 and Aquilegia 022229) while 28 % had no SORLIP1 sequences. A phylogenetic tree of the ELIP sequences showed a broad distribution for the presence of at least one SORLIP1 element. Three ELIP gene clades were found in Eucalyptus grandis, but only one clade contained ELIP promoters with SORLIP1 elements. In Brachypodium distachyon, one clade had 2–3 SORLIP1 elements while the other had 0–1 SORLIP1 elements. It will be interesting to determine if HL induction levels for Eucalyptus and Brachypodium ELIP paralogs correlate with the presence/absence of SORLIP1 elements.
The dSL element plays a role in HL induction, but ELIP genes have been shown to be regulated by other abiotic and biotic stresses (Hruz et al. 2008). To determine if the dSL element is important in modulating an increase in expression in response to these stresses, WT and dSL mutant transgenic lines will need to be exposed to these stresses and GUS and ELIP mRNA levels will need to be quantified to determine if there are significant differences in GUS expression.
A yeast one-hybrid screen was performed to identify dSL interacting partners, and the C-terminal region of Lhca2 was selected at a high frequency (11 of 30 in-frame clones, Supplemental Figure 4a). Lhca2-encoding clones displayed strong activation with the dSL bait, but no activation for the dSL mutated (dSLm) bait (Supplemental Figure 4b). This specificity was surprising since Lhca2 is a light harvesting complex protein associated with PSI and localized to the chloroplast thylakoid membrane. The C-terminal region of Lhca2 has no cryptic DNA binding domains or a nuclear localization sequence (Rost et al. 2003). The high abundance of LHCB and LHCA mRNAs would result in a high abundance of LHC-encoding cDNAs, however if the LHCA2 clones were random selection artifacts, many other LHCB and LHCA cDNAs would have been selected as well: not just one region of one LHC-encoding cDNA. A genetic analysis (ELIP1 and ELIP2 mRNA induction after HL treatment in an lhca2 T-DNA insertion mutant (Alboresi et al. 2009)) did not support a role for Lhca2 in HL induction of ELIP mRNA, and thus the specific activation of the dSL bait by the C-terminal region of Lhca2 observed in yeast does not appear to extend to Arabidopsis.
In conclusion, the double SORLIP1 element has been shown to be required for HL induction of ELIP1 and ELIP2 promoters in A. thaliana using transgenic lines with promoter-reporter constructs. In addition, two G-box elements redundantly contributed to the HL induction of the ELIP genes. ELIP gene promoters throughout the plant kingdom display a small level of enrichment for SORLIP1 elements.
References
Adamska I (1997) ELIPs-light-induced stress proteins. Physiol Plant 100:794–805
Adamska I, Kloppstech K, Ohad I (1993) Early light-inducible protein in pea is stable during light stress but is degraded during recovery at low light intensity. J Biol Chem 268:5438–5444
Adamska I, Roobol-Boza M, Lindahl M, Andersson B (1999) Isolation of pigment-binding early light-inducible proteins from pea. Eur J Biochem 260:453–460
Alboresi A, Ballottari M, Hienerwadel R, Giacometti G, Morosinotto T (2009) Antenna complexes protect photosystem I from photoinhibition. BMC Plant Biol 9:71–84
Arguello-Astorga G, Herera-Estrella L (1998) Evolution of light-regulated plant promoters. Ann Rev Plant Physiol Plant Mol Biol 49:525–555
Blecken J, Weisshaar B, Herzfeld F (1994) Two distinct cis-acting elements are involved in light-dependent activation of the pea elip promoter. Mol Gen Genet 245:371–379
Brusslan J, Peterson M (2002) Tetrapyrrole regulation of nuclear gene expression. Photosynth Res 71:185–194
Butaye K, Cammue B, Delaure S, De Bolle M (2005) Approaches to minimize variation of transgene expression in plants. Mol Breed 16:79–91
Chattopadhyay S, Ang L-H, Puente P, Deng X-W, Wei N (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10:673–683
Clough SJ, Bent AJ (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Duanmu D, Casero D, Dent R, Gallaher S, Yang W, Rockwell N, Martin S, Pellegrini M, Niyogi K, Merchant S et al (2013) Retrograde bilin signaling enables Chlamydomonas greening and phototrophic survival. Proc Natl Acad Sci USA 110:3621–3626
Ensminger I, Sveshnikov D, Campbell D, Funk C, Jansson S, Lloyd J, Shibistova O, Oquist G (2004) Intermittent low temperatures constrain spring recovery of photosynthesis in boreal Scots pine forests. Glob Chang Biol 10:995–1008
Estavillo G, Crisp P, Pornsiriwong W, Wirtz M, Collinge D, Carrie C, Giraud E, Whelan J, David P, Javot H et al (2011) Evidence for a SAl1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23:3992–4012
Gadjev I, Vanderauwera S, Gechev T, Laloi C, Minkov I, Shulaev V, Apel K, Inze D, Mittler R, Van Breusegem F (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141:436–445
Heddad M, Adamska I (2002) The evolution of light stress proteins in photosynthetic organisms. Comp Funct Genomics 3(6):504–510
Heddad M, Noren H, Reiser V, Dunaeva M, Andersson B, Adamska I (2006) Differential expression and localization of early light-induced proteins in Arabidopsis. Plant Physiol 142:75–87
Higo K, Ugawa Y, Iwatmoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27:297–300
Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Wildmayer P, Gruissem W, Zimmermann P (2008) Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinform 2008:1–5. Article ID 420747
Hudson M, Quail P (2003) Identification of promoter motifs involved in the network of phytochrome A-regulated gene expression by combined analysis of genomic sequence and microarray data. Plant Physiol 133:1605–1616
Hutin C, Nussaume L, Moise N, Moya I, Kloppstech K, Havaux M (2003) Early light-induced proteins protect Arabidopsis from photooxidative stress. Proc Natl Acad Sci USA 100:4921–4926
Jefferson R, Kavanagh T, Bevan M (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Kleine T, Leister D (2013) Retrograde signals galore. Frontiers Plant Sci 4:Article 45
Kleine T, Voigt C, Leister D (2009) Plastid signalling to the nucleus: messengers still lost in the mists? Trends Genet 25:185–192
Klimmek F, Sjodin A, Noutsos C, Lester D, Jansson S (2006) Abundantly and rarely expressed Lhc protein genes exhibit distinct regulation patterns in plants. Plant Physiol 140:793–804
Kolanus W, Scharnhorst C, Kuhne U, Herzfeld F (1987) The structure and light-dpendent transient expression of a nuclear-encoded chloroplast protein gene from pea (Pisum sativum L.). Mol Gen Genet 209:234–239
Koussevitzky S, Nott A, Mockler T, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J (2007) Signals from the chloroplasts converge to regulate nuclear gene expression. Science 316:715–719
Kuhlemeier C, Green P, Chua N-H (1987) Regulation of gene expression in higher plants. Ann Rev Plant Physiol 38:221–257
Leon P, Gregorio J, Cordoba E (2013) ABI4 and its role in chloroplast retrograde regulation. Fronteirs Plant Sci 3:304
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Maddison W, Maddison D (2011) Mesquite: a modular system for evolutionary analysis. Version 2.75:http://mesquiteproject.org
Mann H, Whitney D (1947) On a test of whether one of two random variables is stochastically larger than the other. Ann Math Stat 18:50–60
Meyer G, Kloppstech K (1984) A rapidly light-induced chloroplast protein with a high turnover coded for by pea nuclear DNA. Eur J Biochem 138:201–207
Morey C, Mookherjee S, Rajasekaran G, Bansal M (2011) DNA free energy-based promoter prediction and comparative analysis of Arabidopsis and rice genomes. Plant Physiol 156:1300–1315
op den Camp R, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg E, Gobel C, Feussner I et al (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15:2320–2332
Rossel J, Wilson P, Hussain D, Woo N, Gordon M, Mewett O, Howell K, Whelan J, Kazan K, Pogson B (2007) Systemic and intracellular responses to photooxidative stress in Arabidopsis. Plant Cell 19:4091–4110
Rossini S, Casazza A, Engelmann E, Havaux M, Jennings R, Soave C (2006) Suppression of both ELIP1 and ELIP2 in Arabidopsis does not affect tolerance to photoinhibition and photooxidative stress. Plant Physiol 141:1264–1273
Rost B, Yachdav G, Liu J (2003) The predictprotein server. Nucleic Acids Res 32:W321–W326
Tepperman J, Zhu T, Chang H-S, Quail P (2001) Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA 98:9437–9442
Tzvetkova-Chevolleau T, Franck F, Alawady A, Dall’Osto L, Carriere F, Bassi R, Grimm B, Nussaume L, Havaux M (2007) The light stress-induced protein ELIP2 is a regulator of chlorophyll synthesis in Arabidopsis thaliana. Plant J 50:795–809
Van Aken O, Whelan J (2012) Comparison of transcriptional changes to chloroplast and mitochondrial perturbations reveals common and specific responses in Arabidopsis. Froneris Plant Sci 3:281
Acknowledgments
We thank Dr. Simon Malcomber for his help with the Mesquite analysis and Dr. James Archie for his statistics expertise.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Rus Alvarez-Canterbury, A.M., Flores, D.J., Keymanesh, K. et al. A double SORLIP1 element is required for high light induction of ELIP genes in Arabidopsis thaliana . Plant Mol Biol 84, 259–267 (2014). https://doi.org/10.1007/s11103-013-0130-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-013-0130-4