Abstract
In plants, RNA-directed DNA methylation (RdDM) and related transcriptional gene silencing (TGS) involve members of the suppressor of variegation 3-9-homologous (SUVH) group of putative histone methyltransferases. Utilizing a reverse genetic approach in Arabidopsis thaliana, we demonstrate that two closely related SUVH members, SUVH2 and SUVH9, act partially non-redundant in RdDM. DNA methylation, transcript accumulation and association with histone modifications were analyzed at the endogenous RdDM target AtSN1 (a SINE-like retroelement) in suvh2 and suvh9 single as well as suvh2 suvh9 double mutants. SUVH2 was found to be required for full DNA methylation at AtSN1 in early seed development and was also higher expressed in seeds than at later developmental stages. SUVH9 had its impact on RdDM later during vegetative development of the plant and was also higher expressed during that stage than at earlier developmental stages. The strongest reduction of RdDM at AtSN1 was found in suvh2 suvh9 double mutant plants. Histone 3-lysine 9-dimethylation (H3K9me2) associated with AtSN1 was reduced only in the simultaneous absence of functional SUVH2 and SUVH9. Thus, SUVH2 and SUVH9 functions in RdDM and TGS are overlapping in spite of some developmental specialization. Pol V specific transcripts were reduced in suvh2 suvh9 plants. This might indicate a role of these SUVH proteins in Pol V complex recruitment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
SET domain containing proteins can be divided into four different classes typified by the corresponding proteins of Drosophila E(Z), TRX, ASH1 and SU(VAR)3-9 (Baumbusch et al. 2001; Thorstensen et al. 2011). In A. thaliana, 15 genes similar to the SU(VAR)3-9 were identified, of which 10 belong to the group of suppressor of variegation [SU(VAR)3-9] homologous genes (SUVH) defined by the presence of an internal YDG/SRA-domain and C-terminal pre-SET and SET domains. Among those, SUVH2 (encoded by At2g33290) and SUVH9 (encoded by At4g13460), show considerable sequence similarity (Naumann et al. 2005) and form a subgroup within the SUVH-family that is characterized by the absence of the catalytically important post-SET domain (Rea et al. 2000). SUVH2 has been reported to participate in gene silencing via heterochromatization (Naumann et al. 2005; Ay et al. 2009) and both, SUVH2 and SUVH9, are involved in DRM2-mediated de novo DNA methylation of particular target regions in A. thaliana (Johnson et al. 2008). The SET domain is essential for histone methyltransferase activity (Rea et al. 2000). Nevertheless, in vitro histone methylation tests were successful so far only for a subset of SUVH proteins, possibly due to rather specific substrate requirements of some of the family members. SUVH4, SUVH5 and SUVH6 readily methylate bovine H3 (Jackson et al. 2004; Ebbs and Bender 2006). SUVH5 additionally showed some activity on bovine H2A (Ebbs and Bender 2006). SUVH2 has been reported to methylate H4 in one study using reconstituted recombinant nucleosomes (Naumann et al. 2005), but was found inactive in a second study using bovine histones or A. thaliana nucleosomes (Johnson et al. 2008).
The second characterized domain in the SUVH proteins is the SET and RING associated (SRA) domain (Johnson et al. 2007, also termed YDG domain according to Baumbusch et al. 2001). This domain was shown to be responsible for the binding of proteins to methylated DNA in vitro (Rajakumara et al. 2011). The SRA domains of histone methyltransferases SUVH4 and SUVH6, which are linked to MET1- and CMT3-mediated maintenance of DNA methylation, have preferential affinity to methylated cytosines in CHG and CHH (H indicating A,C or T) context (Johnson et al. 2007). SUVH2 and SUVH9 are involved in de novo RNA-directed DNA methylation (RdDM) (Naumann et al. 2005; Johnson et al. 2008). The SRA domain of SUVH2 shows high affinity to methylated cytosines in CG motifs, while the SRA domain SUVH9 has the strongest affinity to methylated cytosines in CHH context (Johnson et al. 2008). Based on these results, Johnson et al. (2008) postulated that SUVH2 and SUVH9 may act in RdDM rather via binding to methylated cytosines in particular sequence contexts, than by actually methylating histones.
AtSN1 (A. thaliana SINE like element 1) is a putative transposable retroelement present in 71 copies in the A. thaliana Col-0 reference genome (Myouga et al. 2001). A single copy of AtSN1 located on chromosome 3 that is flanked by genic sequences has served as a model for the genetic analysis of RdDM of genomic sequences of A. thaliana in many studies (Zilberman et al. 2003; Xie et al. 2004; He et al. 2009a, b; Gao et al. 2010). Its internal region between the long terminal repeats (LTR) contains in total 44 cytosine sites; 11 in symmetric (4 CG sites, 7 CHG sites) and 33 in asymmetric (CHH) context. ~40 % of these cytosines are methylated in wild type plants. In symmetric context, cytosine methylation of AtSN1 and other plant sequences, is primarily mediated by the DNA methyltransferase MET1 (Finnegan et al. 1993; Lindroth et al. 2001; Bartee et al. 2001) while methylation in asymmetric context is mainly mediated by DRM2 (RdDM pathway; Cao and Jacobsen 2002). In addition to the de novo DNA methyltransferase DRM2, the DNA dependent RNA polymerases Pol VI and Pol V contribute to RdDM and transcriptional suppression of AtSN1 (Wierzbicki et al. 2008) by synthesis of siRNA homologous to the methylated region (Pontier et al. 2005). Further factors involved in RdDM are RDR2, DRD1 and DDM1 (Wierzbicki et al. 2009). Most likely, the RdDM pathway involves a self amplifying feedback loop of siRNA dependent DNA methylation and DNA methylation dependent Pol IV-RDR2 mediated formation of siRNAs (Law and Jacobsen 2010).
In wild type plants with functional RdDM, no AtSN1 sense transcripts are found (Wierzbicki et al. 2009; Haag et al. 2009), although the internal LTR-flanking region of AtSN1 shares structural similarity with promoters of tRNA (Box A and Box B) and of genes typically transcribed by DNA-dependent RNA Pol III (Myouga et al. 2001). Formation of AtSN1 sense transcripts is seen in mutants with disturbed RdDM such as nrpe1, ago4 and dms3. As the transcriptional activation was not associated with acetylation of H3 at lysines 9 and 14 or Pol II association at AtSN1, it is assumed that AtSN1 transcription is Pol III-dependent (Wierzbicki et al. 2009). AtSN1 silencing in wild type plants is associated with complementary and adjacent antisense transcripts, which are most likely products of Pol V (Wierzbicki et al. 2008) which are missing in nrpe1 mutant plants (NRPE1 is the largest subunit of the Pol V complex) and reduced in rdm3 mutants (He et al. 2009a, b, c). Interaction of NRPE1 with the AtSN1 region was shown by chromatin immunoprecipitation (ChIP) (Rowley et al. 2011).
In the silenced state, the AtSN1 region is associated with high levels of repressive histone marks such as H3K9me2 and H3K27me1, while acetylation of histone 3 is mainly absent (Wierzbicki et al. 2008). Addionally, association of AtSN1 with H3K4me2, a mark associated with active chromatin was described, defining this region as “intermediate heterochromatin” (Habu et al. 2006). In plants without functional Pol V, the level of H3K9me2 remained unaltered in the internal AtSN1 region (Wierzbicki et al. 2008).
Here, we show that the two putative histone methyltransferases SUVH2 and SUVH9 are differentially expressed during plant development and contribute, at least in part, non-redundantly to RdDM. SUVH2 plays a major role at the seedling stage and SUVH9 during vegetative growth. Nevertheless, both are important for transcriptional silencing and persistence of repressive histone marks.
Materials and methods
Plant material and growth conditions
The A. thaliana accession Col-0 was used in all experiments. The suvh2 T-DNA insertion lines were ordered from NASC (Nottingham Arabidopsis Stock Centre, T-DNA insertion line Gabi_kat_516A07, Suppl. Figure 2). The suvh9 T-DNA insertion line was ordered from NASC (T-DNA insertion line SALK_048033). Double mutant suvh2 suvh9 line was generated by crossing Gabi_kat_516A07 and SALK_048033 and genotyping of the F3 generation. SUVH2 over-expression plants were obtained from the laboratory of G. Reuter (Martin-Luther University Halle-Wittenberg, Germany). These plants contain a strong constitutive expressed myc-tagged SUVH2 gene controlled by the CaMV35 promoter. Plants with enhanced level of SUVH2 were selected by their dwarf phenotype, curled leaves and delayed senescence.
Plants were cultivated on soil at 21 °C under a 16 h light/8 h dark (long day) regime for propagation and seed production or under an 8 h light/16 h dark (short day) regime to generate material for RNA and DNA analysis. All seeds were stratified before germination for 3 days at 4 °C in the dark. Kanamycin and phosphinotricin (BASTA®) resistance of the plants was tested on germination medium (1/2 MS salts; 10 g/l sucrose) plates with 40 mg/l kanamycin or 20 mg/l Glufosinate-ammonium under long day conditions.
Primers used for genotyping: Supplementary Table.
RNA analysis
Total RNA samples for transcript measurements were extracted from seedlings grown for 1 week on GM medium under long day conditions and from rosette leaves of plants grown for 6–8 weeks under short day conditions. For analysis of RNA in imbibed seeds, seeds were soaked in water over night at RT. RNA was prepared in triplicates from 100 mg of seedlings grown on agarplates. Total RNA and small RNA were extracted from 1 g tissue with the mirVana™ miRNA Isolation kit (Ambion) according to manufacturers protocol E. and F. II. cDNA was prepared using RevertAid™ H Minus M-MuLVRT (Fermentas) according to manufacturers protocol. mRNA and AtSN1-A analysis was performed on oligo dT transcribed cDNA, AtSN1-B (and C) on cDNA using specific primers. cDNAs were quantified by the RT-qPCR method using the iCycler (Bio-rad) and the iQ™ SyBR® Green Supermix (Bio-rad). Program: 1: 5′ 95 °C; 2: 15″ 95 °C; 3: 30″ 65 °C; 4: 30″ 72 °C; 5: goto2 40×; 6: Melting curve 65 °C 10″ +0.5 °C 60 repeats; 7: 4 °C.
Primers used for qPCR are defined in the Supplementary Table.
For each genotype, 5 individual plants were analyzed. Mid values are indicated as bar heights, standard deviation of values as error bars (Fig. 1, Suppl. Figure 3).
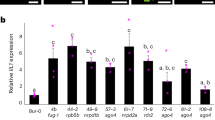
Differential expression of SUVH2 and SUVH9 in wild type A. thaliana. SUVH2 (yellow) and SUVH9 (orange) mRNAs were quantified relative to ACT2 mRNA at 3 different developmental stages using RT-qPCR: Imbibed seeds: 24 h imbibing of the seeds in water, Seedlings: grown for 7 days on agar plates after stratification, and Leaves: leave tissue from plants grown for 6 weeks on soil. The depicted bars give the median and the error bars the total deviation of 5 independent biological samples
ACTIN2 (ACT2; At3g18780) and PHOSPHOFRUCTOKINASE (PFK; At4g04040; transcripts only present in seedlings and leaf tissue) were used as reference genes (Correlation Coefficient and PCR efficiency: Suppl. Figure 8).
Northern blot analysis of siRNA/miRNA
Small RNA were detected by Northern blot analysis of a RNA fraction (10 μg per lane for miRNA 157a and 50 μg per lane for AtSN1 siRNA) enriched for small molecules. Hybridization was performed as described by Mette et al. (2005) using a 32P-labeled AtSN1 antisense transcript.
DNA methylation analysis (Bisulfite sequencing)
Genomic DNA for methylation analysis was extracted with a DNeasy Plant Maxi Kit (Qiagen, Hilden, Germany) from rosette leaves of individually genotyped plants grown for 6–8 weeks under short day conditions and from seedlings grown on GM medium for 1 week under long day conditions.
Bisulfite-mediated chemical conversion of DNA was done using a Qiagen EpiTect Bisulfite Kit (Qiagen, Hilden, Germany) following the manufacturers instructions. Amplicons were transformed with the help of the StrataClone™ PCR Cloning Kit. Sequencing of single clonal colonies was done with the Cyclereader Auto Sequencing Kit (Fermentas, St.Leon-Rot, Germany) and Licor 4300 DNA Analyzer.
Statistical analysis was performed using Chi-square test comparing the number of methylation events out of all possible cytosine sites detected by bisulfite sequencing. Standard deviation is calculated by QUICKBASIC Program for Exact and Mid-P Confidence Intervals for a Binomial Proportion (see Suppl. Fig 4a–h).
Primers used for amplification of converted target regions are defined in the Supplementary Table.
Chromatin immuno precipitation (ChIP)
The ChIP was performed according Jovtchev et al. (2011), Gendrel et al. (2005) and Lawrence and Pikaard (2004) with modifications. Four g of 1 week old seedlings were used for crosslinking by 1 % w/v formaldehyde solution under vacuum for 15 min (seedlings) or 10 min (leaves). Crosslinking was quenched with 2.5 ml 2 M glycine under vacuum for five min. The plant material was homogenized in liquid nitrogen and suspended in 30 ml extraction buffer. The extract was filtered through 2 layers of Miracloth and centrifuged at 1,800g for 20 min at 4 °C. The pellet was resuspended in 1 ml of extraction buffer 2 and centrifuged at 16,000g for 10 min. The pellet was resuspended in 300 μl extraction buffer 3 and overlayed on 300 μl of extraction buffer 3. The nuclei are collected by centrifugation at 16,000g for 1 h. The pellet was resuspended in 300 μl of cold nuclei lysis buffer. The sonication of the chromatin was performed using a Diagenode Disruptor for 6 cycles with 30 s of high energy sonication and 30 s break at 4 °C. Nuclear debris was removed by centrifugation at 16,000g for 10 min. One aliquot of the chromatin extract was used for the input and the others were diluted 1:10 in ChIP dilution buffer. The immunoprecipitation was performed with Dynabead Protein A (Dynal) coupled antibodies. H3: #39163 Histone H3 C-terminal rabbit pAB, Activemotif; H3K4me3: #39159 Histone H3 trimethyl Lys4 Rabbit pAB, Activemotif; H3K9me2: mouse monoclonal [mAbcam1220), Abcam; H3K27me3:; H4K20me1: ab9051, Rabbit polyclonal Histone H4 (monomethyl K20), Abcam were used.
After overnight incubation at 4 °C, the coupled beads were washed 2 times 5 min each with low salt buffer :150 mM NaCL, 0.1 % SDS, 1 % Triton X100, 2 mM EDTA, 20 mM Tris–HCl (pH 8.1); high salt buffer: 500 mM NaCL, 0.1 % SDS, 1 % Triton X-100, 2 mM EDTA, 20 mM Tris–HCl (pH 8.1); LiCl wash buffer: 0.25 LiCl, 1 % NP40, 1 % Na deoxycholate, 1 mM EDTA, 10 mM Tris–HCl (pH 8.1) and TE buffer: 10 mM Tris–HCl (pH 8.), 1 mM EDTA. The DNA was eluted twice by incubation with 250 μl elution buffer at 65 °C for 15 min.
Primers used for quantitative amplification of genomic regions after ChIP are defined in the supplementary table.
Results
SUVH2 and SUVH9 are differentially expressed during A. thaliana development
Transcripts of SET-domain protein family genes SUVH2 (At2g33290) and SUVH9 (At4g13460) are detectable throughout the lifecycle of A. thaliana, but show different expression levels when material of different stages of plant development is compared (Suppl. Figure 1). In seeds imbibed in water for 1 day, SUVH2 expression was highest and decreased during seedling development and in mature leaves of 6 week old plants grown under short day conditions (Fig. 1). SUVH9 showed an expression pattern inverse to that of SUVH2, i.e., a low SUVH9 transcript abundance in seeds and a higher one in seedlings and leaves of mature plants. The observed changes in SUVH2 and SUVH9 expression could be due to differential gene regulation over developmental time or, provided that SUVH2 and SUVH9 are preferentially expressed in different cell types, might reflect the different cell type-composition of the analysed tissues.
Single suvh2 and suvh9 mutations affect RdDM to different extent at different developmental stages
To test the significance of this differential expression of SUVH2 and SUVH9, we applied a reverse genetic approach. T-DNA segregation analysis, sequencing of the PCR-amplified T-DNA integration locus (Suppl. Figure 2) and transcript quantification by RT-qPCR (Suppl. Figure 3) in T-DNA insertion lines identified the loss-of-function alleles suvh2 (GABI-Kat 516A07; Rosso et al. 2003) and suvh9 (SALK_048033; Alonso et al. 2003; Johnson et al. 2008) that both contained a single T-DNA insertion locus and showed robust specific reduction of SUVH2 and SUVH9 transcripts, respectively. A suvh2 suvh9 double mutant line was obtained by crossing of the single mutant lines followed by self-pollination of F1 and F2 plants. Homozygous suvh2 suvh9 plants were identified in the F3 generation using PCR-based markers selective for T-DNA insertion and wild type alleles. For all following analyses, material from homozygous F4 and F5 plants was used.
The dispersed repetitive element AtSN1 is known to undergo persistent RNA-directed DNA methylation and transcriptional silencing in wild type A. thaliana Col-0 throughout almost the whole life cycle of the plant. A particular copy of AtSN1 located on chromosome 3 between annotated genes At3g44000 and At3g44005 (Fig. 2), nucleotides 15,805,617–15,805,773 of chromosome 3 (NCBI nucleotide sequence NC003074), has been used in several studies to check the effect of mutations on RdDM at endogenous sequences (Wierzbicki et al. 2008, 2009; Johnson et al. 2008). DNA methylation at this copy was determined by bisulfite sequencing of genomic DNA from seedlings and leaves of mature 6 week old wild type plants, suvh2 and suvh9 single mutant as well as suvh2 suvh9 double mutant lines (Fig. 3a, Supplementary Fig. 4A–H). Methylation levels were calculated in percent methylated cytosines of total cytosines, cytosines in CG, in CHG and in CHH context. In wild type plants, AtSN1 DNA methylation was essentially the same in seedlings and leaves. Methylation was strongest reduced in suvh2 suvh9 double mutants. This reduction affected cytosines in all contexts and was rather similar in seedlings and leaves from mature plants. Thus, SUVH2 and SUVH9 have redundant roles, as the double mutant line displays a more pronounced effect on RdDM than the single mutants. Nevertheless, in seedlings, the suvh2 mutation caused a stronger reduction of total DNA methylation than the suvh9 mutation. This seemed mainly due to a stronger effect of suvh2 on CHH context methylation. In contrast, a stronger reduction of total DNA methylation in the suvh9 compared to the suvh2 was found for leaves from mature plants. During development of seedling to mature plants, the level of DNA methylation in the suvh9 mutant is consistently reduced, while it remains constant in the suvh2. Consistent with the peaks of SUVH2 and SUVH9 transcripts in seedlings and leaves of mature plants, respectively, SUVH2 seems to be more important for RdDM in seedlings and SUVH9 in mature leaves. Thus, SUVH2 and SUVH9 act at least in part developmentally non-redundantly over the plant life cycle.
AtSN1 as a model target of RNA-directed DNA methylation and transcriptional silencing. Schematic overview for the genomic region including SINE-like element AtSN1 located on chromosome 3 next to At3G44005. LTRs (bold grey arrows) flank the inner region (bold black arrow) that contains two putative promoter-like structures, Box A and Box B, and the internal region with homology to tRNA genes. Black dots indicate the region in which methylated cytosines are detected in wild type. Positions of primers used for amplification after bisulfite conversion bs-f and bs-r indicate the region analyzed for DNA methylation. The dotted arrow shows the putative extent of silenced transcript A, the black arrows indicate Pol V derived transcripts B and C. Positions of primers used for RT-PCR are marked by A-f, A-r; B-f, B-r; C-f, C-r
AtSN1 and AtCOPIA4 DNA methylation in suvh2, suvh9 and suvh2 suvh9 mutants. DNA methylation was determined by bisulfite sequencing of genomic DNA from 7 day old seedlings or leaves of mature 6 week old plants. Individual clones (N) were sequenced for the different genotypes. The bars mark the levels of DNA methylation in percent of methylated cytosines relative to total cytosines, with black indicating all cytosines, dark grey CG context, light grey CHG context and red indicating CHH context. Wt indicates wild type plants. a DNA methylation in AtSN1. Significance of differences in total DNA methylation between the different genotypes were checked by Chi Square tests (table, ***p < 0.001 and ANOVA, data not shown). b DNA methylation at AtCOPIA4
AtSN1 methylation was also determined in leaves from a line hemizygous for a Pro35S-mycSUVH2 over-expression construct (Naumann et al. 2005; Ay et al. 2009). Despite their dwarf habitus typical for increased SUVH2 transcript levels, Pro35S-mycSUVH2 plants did not show an obvious increase in AtSN1 methylation (Suppl. Figure 5). Thus, SUVH2 is required for RdDM of AtSN1, but is not the rate limiting factor if functional SUVH2 and SUVH9 alleles are present.
The GC-rich retroelement AtCOPIA4 (At4g16870) is hypermethylated in the A. thaliana genome, mainly due to maintenance methylation involving MET1 and SUVH4 (Johnson et al. 2007). AtCOPIA4 methylation is hardly affected in mutants of RdDM components. We performed bisulfite sequencing of an AtCOPIA4 element containing 83 cytosines (14 in CG, 6 in CHG and 63 in CHH context) (Fig. 3b, Suppl. Figure 4 I to P) in order to test for effects suvh2 and suvh9 mutations on its DNA methylation. No obvious changes of DNA methylation were found for all genotypes in seedlings and leaves from mature plants indicating that COPIA4 methylation is apparently not affected by mutants of the RdDM pathway.
Only the suvh2 suvh9 double mutant shows released silencing of AtSN1 sense transcription
The AtSN1 region is the origin of three different transcripts (Fig. 2). In wild type plants, the AtSN1 region is silenced and Pol III dependent transcription of sense transcript (A) is suppressed, whereas Pol V dependent antisense transcripts (B and C) are detectable (Wierzbicki et al. 2008). Thus, sense transcript A and antisense transcripts B and C were supposed to be mutually exclusive (Wierzbicki et al. 2008). We determined AtSN1 transcripts A and B by reverse transcription combined with ACT2 mRNA in RNA extracted from imbibed seeds and leaves of 6 week old wild type plants, suvh2, suvh9 and suvh2 suvh9 double mutant lines (Fig. 4a). Consistent with Wierzbicki et al. 2008, the level of transcript A was low and that of transcript B was high in wild type seeds and leaves. The inverse pattern was found for the suvh2 suvh9 double mutant, where transcript A was increased and transcript B was reduced. The results for suvh2 and suvh9 single mutants were less clear. The increase of transcript A was not prominent, and transcript B was somewhat reduced in imbibed seeds. The effects of mutations in suvh2 and suvh9 on AtSN1 transcript A levels were confirmed by reverse transcription combined with quantitative PCR (RT-qPCR; Fig. 4b). In comparison to wild type, AtSN1 transcript A levels all in suvh2 suvh9 double mutants were increased 14 fold in imbibed seeds, 6 fold in seedlings and threefold in leaves of mature plants. Thus, with regard to their effects on AtSN1 transcripts, SUVH2 and SUVH9 seem to act redundantly. The release of silencing of AtSN1 in suvh2 suvh9 double mutants was associated with a decrease of siRNAs that targets AtSN1 (Suppl. Figure 6).
AtSN1-derived transcripts in suvh2, suvh9 and suvh2 suvh9 mutants. AtSN1 transcripts were determined relative to ACT2 and PFK mRNA in total RNA from imbibed seeds, 7 day old seedlings or leaves of mature 6 week old plants by RT PCR-based methods with primers specific for transcripts A and B. a Semiquantitative PCR analysis of AtSN1 transcripts A and B in imbibed seeds and leaves in comparison to ACT2 mRNA. b RT-qPCR analysis of AtSN1 transcript A (yellow) and PFK mRNA (grey) in imbibed seeds, leaves and seedlings relative to ACT2 mRNA. The bars indicate the median of 5 biological samples measured in technical duplicate; the error bars indicate the total deviation among samples
To investigate the involvement of SUVH2 in silencing of Pol II dependent transcription, we used of a well characterized two component transgene system mediating RNA-directed transcriptional gene silencing (TGS; Aufsatz et al. 2002; Fischer et al. 2008). Although impaired reporter gene silencing was seen in the suvh2 mutant (Suppl. Figure 7), this observation turned out to be inconclusive due to the structure of the used suvh2 T-DNA insertion allele. Line GABI-Kat 516A07 contains two T-DNA copies in a tail-to-tail arrangement (Suppl. Figure 2). The resulting inverted repeat structure of CaMV Pro35S sequences leads to TGS of the silencer Pro35S-NOSpro-proNOS construct. Such cross-silencing effects were already reported for other T-DNA insertion lines (Daxinger et al. 2008).
Release of AtSN1 sense transcription in suvh2 suvh9 is correlated with altered histone modification
DNA hypermethylation and transcriptional silencing are usually associated with increased nucleosome density and histone modifications typical for repressed chromatin such as H3K9me2 (Pecinka et al. 2010). Thus, chromatin immunoprecipitation combined with quantitative PCR (ChIP qPCR) was performed to quantify H3K4me3 and H3K9me2 associated with different regions of AtSN1 in 7 day old seedlings (Fig. 5). To determine nucleosome density, we used an antibody specific for H3 irrespective of modifications. Primers were designed to separately analyze AtSN1 regions corresponding to sense transcript A and to antisense transcripts B and C (Fig. 2). Wild type values were set to 1.0, and values for mutants were calibrated accordingly. As controls, the highly transcribed gene PFK typical for active and the retroelement Ta3 typical for inactive chromatin were used (Johnson et al. 2002; Mathieu et al. 2003, 2005). As PFK shows very little association with H3K9me2, ChIP-qPCR for this mark at PFK is in particular error-prone. After calculation of relative values for mutants in comparison to wild type, this high initial errors result in a high deviation of values. The analysis of H3 density at AtSN1, PFK and Ta3 did not show consistent differences between wild type and mutant plants that would indicate altered nucleosome density at these sequences in suvh2 or suvh9 mutants. If at all, a tendency for lower nucleosome density at AtSN1 might be seen in the suvh2 suvh9 double mutant, which would be concordant with the increased transcriptional activity. H3K4me3 as a histone mark associated with actively transcribed chromatin (Fuchs et al. 2006; Moghaddam et al. 2011) was slightly increased in the suvh2 suvh9 double mutant at the AtSN1 region homologous to transcript A. No consistent change was seen, neither for other sequences, nor in single suvh2 and suvh9 mutants. H3K9me2 is typical for transcriptional inactive chromatin (Stancheva 2005). In the suvh2 suvh9 double mutant, it was found reduced in the regions A, B and C of AtSN1. The reduction of H3K9me2 was more evident than the reduction in nucleosome density, indicating that the measurements reflected a genuine reduction of H3K9me2 rather than a reduction of nucleosome density. Again, no consistent changes were seen for other sequences analyzed, and also not in single suvh2 and suvh9 mutants. Thus, with regard to their effects on H3K4me3 and H3K9me2 density at transcribed AtSN1 regions, SUVH2 and SUVH9 seem to act mainly redundantly. Thus, SUVH2 and SUVH9 non-redundancy seems limited to their role in DNA methylation. H3K27me2 and H4K20me1 were also analyzed at AtSN1, but no obvious changes were found between mutants and the wild type (data not shown).
AtSN1-associated histone modifications in suvh2, suvh9 and suvh2 suvh9 mutants. Chromatin immunoprecipitation combined with quantitative PCR (ChIP-qPCR) was performed on 1 week old seedling with antibodies specific to H3, H3K4me3 and H3K9me2 and primers specific for PFK, AtSN1 regions A, B, C and Ta3. Wild type values were set to 1.0 and values for mutants were calibrated accordingly. Black indicates wild type, light-grey suvh2 single mutants, dark-grey suvh9 single mutants and red suvh2 suvh9 double mutants. Quantification by qPCR was performed in duplicates. Bars indicate the mean; error bars the standard deviation from 3 independent experiments. a H3, b H3K4me3, c H3K9me2
Discussion
During plant reproduction, processes of genome-wide epigenetic reprogramming are taking place (Slotkin et al. 2009). This can include changes of histone modifications, DNA methylation and siRNA levels. Nevertheless, potentially harmful elements such as AtSN1 stay transcriptional silenced throughout development. Both SET domain proteins SUVH2 and SUVH9 are required for complete transcriptional silencing of AtSN1, acting in a seemingly redundant way. However, with regard to RNA-directed DNA methylation, SUVH2 and SUVH9 activities are at least in part developmentally non-redundant. This might be related to the different binding properties of the two proteins for methylated DNA (Johnson et al. 2008). Taking into account the reported in vitro binding preferences of the SUVH2 (methylated cytosines in CG context) and SUVH9 (methylated cytosines in CHH context) and the ratio of potentially methylated cytosines in these contexts in AtSN1 (4 sites in CG vs. 33 CHH context), one would predict that loss of SUVH9 should have a stronger impact than loss of SUVH2, irrespective of the developmental stage of sampled plants. The effects of the single suvh2 and suvh9 mutations on AtSN1 methylation levels observed in our study do not meet this expectation. Rather, from the observed reduction of DNA methylation in suvh2 seedlings and suvh9 mature leaves, we would conclude that the differential expression patterns of SUVH2 and SUVH9 can explain their non-redundant involvement in RdDM over development. The high expression of the SUVH2 in imbibed seeds indicates a possible role in the epigenetic reprogramming during early development. Stage-specific regulation of SUVH2 might explain the delayed senescence phenotype observed by Ay et al. (2009) due to ectopic SUVH2 expression during late development and the resulting mis-regulation of senescence controlling genes.
In A. thaliana interphase nuclei, most heterochromatin resides in structures called chromocenters, which are characterized by highly methylated DNA and specific marks such as histone H3K9me2 on the molecular level (Soppe et al. 2002). A clear reduction of histone H3K9me2 at chromocenters has been reported for example for a suvh4 suvh5 suvh6 triple mutant (Johnson et al. 2008; Jovtchev et al. 2011). For a suvh2 single mutant a reduction of H3K9me2 at chromocenters of young leaves has been reported (Naumann et al. 2005), while for the same allele in a suvh2 suvh9 double mutant no immunologically detectable change of H3K9me2 in chromocenters was reported in another study (Johnson et al. 2008). This might be explained by different availability of SUVH2 or SUVH9 at different developmental stages as well as by possible intensity changes of the histone marks themselves during development. While for example chromocenters show highly condensed H3K9me2 and H3K27me2 signals in nuclei of mature leaves, these signals become successively decondensed in nuclei of senescent leaves (Ay et al. 2009).
SUVH2 and SUVH9 are required for proper regulation of RdDM targets such as AtSN1 (Fig. 6). When SUVH2 and SUVH9 were absent, only the AtSN1 region covered by sense transcript A showed an increased association with H3K4me3 concurrent with transcriptional activation. The regions covered by antisense transcripts B and C did not show such a shift to active marks and Pol V-dependent B transcript was not observed. Nevertheless, the repressive mark H3K9me2 was reduced over the entire AtSN1 sequence. How does this coordinated loss of H3K9me2 happen? We assume that the size of the analyzed AtSN1 region plays an important role. As the size of the region corresponding to transcript A correlates approximately to one nucleosome, compact and “locked” structures involving several nucleosomes cannot be established. Such lack of condensation might keep the AtSN1 region in the state of intermediate heterochromatin (Habu et al. 2006). Such intermediate heterochromatin could provide the basis for SET domain protein involvement in silencing. In the absence of SUVH2 and SUVH9, recruitment of the Pol V complex might be impaired, either by the lack of the SET domain proteins themselves with their ability to bind to methylated DNA, or by the lack of a not yet identified histone modification mediated by SUVH2 and SUVH9. The absence of the SET domain proteins or the histone modification mediated by them could then lead to released TGS at the AtSN1 region A accompanied by loss of H3K9me2 and DNA methylation. The absence of Pol V might also allow the recruitment of Pol III resulting in formation of transcript A. The specific association of H3K4me3 with the region covered by transcript A within the analyzed region that lost H3K9me2 implies a hierarchy of these modifications. H3K4me3 modification, associated with active transcription, can only be set when the silencing H3K9me2 is absent. This model is supported by the alteration of histone modifications at the AtSN1 locus observed in suvh2 suvh9 mutants which was not described for Pol V deficient plants (Wierzbicki et al. 2008). In addition, suvh9 and, to a lesser extent, suvh2 single mutants, did not release TGS of the A region despite a significant loss of DNA methylation. Silencing was released only in the suvh2 suvh9 double mutant. Thus, SUVH2 and SUVH9 might act via a pathway independent of Pol V and subsequent de novo DNA methylation. We speculate that SUVH9 and SUVH2 might act via a not yet identified histone methyltransferase activity setting a mark required for subsequent methylation of H3K9 and transcriptional silencing of AtSN1.
Model for SUVH2 and SUVH9 mediated regulation of AtSN1. Schematic overview of the epigenetic status of AtSN1 in wild type plants (upper part) and suvh2 suvh9 mutant plants (lower part) based on the present study. In the wild type (Wt) the AtSN1 region (bold black arrow, with grey arrows indicating the LTR borders) is in a transcriptional silenced state in the presence of both histone methyltransferases SUVH2 and SUVH9. The region undergoing RdDM shows the typical hallmarks of TGS, the presence of AtSN1 A derived 24nt siRNA (red) and DNA methylation (black dots). The AtSN1 derived, Pol III dependent transcript A (dotted arrow) is silenced, while Pol V dependent transcripts B and C (black arrows) can be detected. The transcript A is covering the internal region and the boxes A and B). The most prominent detected histone modification associated with the complete analyzed AtSN1 region is histone H3K9me2 (grey bar). In the absence of SUVH2 and SUVH9 (suvh2 suvh9), silencing is released and the hall marks of RdDM siRNAs and DNA methylation are lost. Pol V dependent transcripts B and C (dotted arrows) are not detectable anymore, while the silencing of transcript A (RNA A black arrow) is released. The complete AtSN1 region looses the association with histone H3K9me2, while the region of active transcription (homolog to RNA A) gets associated with H3K4me3
References
Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Guilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301:653–657
An YQ, McDowell JM, Huang S, McKinney EC, Chambliss S, Meagher RB (1996) Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J 10:107–121
Aufsatz W, Mette MF, van der WJ, Matzke AJ, Matzke M (2002) RNA-directed DNA methylation in Arabidopsis. Proc Natl Acad Sci USA 99(Suppl 4):16499–16506
Ay N, Irmler K, Fischer A, Uhlemann R, Reuter G, Humbeck K (2009) Epigenetic programming via histone methylation at WRKY53 controls leaf senescence in Arabidopsis thaliana. Plant J 58:333–346
Bartee L, Malagnac F, Bender J (2001) Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev 15:1753–1758
Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB (2001) The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res 29:4319–4333
Cao X, Jacobsen SE (2002) Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA 99(Suppl 4):16491–16498
Daxinger L, Hunter B, Sheikh M, Jauvion V, Gasciolli V, Vaucheret H, Matzke M, Furner I (2008) Unexpected silencing effects from T-DNA tags in Arabidopsis. Trends Plant Sci 13:4–6
Ebbs ML, Bender J (2006) Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell 18:1166–1176
Finnegan EJ, Brettell RI, Dennis ES (1993) The role of DNA methylation in the regulation of plant gene expression. EXS 64:218–261
Fischer A, Hofmann I, Naumann K, Reuter G (2006) Heterochromatin proteins and the control of heterochromatic gene silencing in Arabidopsis. J Plant Physiol 163:358–368
Fischer U, Kuhlmann M, Pecinka A, Schmidt R, Mette MF (2008) Local DNA features affect RNA-directed transcriptional gene silencing and DNA methylation. Plant J 53:1–10
Fuchs J, Demidov D, Houben A, Schubert I (2006) Chromosomal histone modification patterns—from conservation to diversity. Trends Plant Sci 11:199–208
Gao Z, Liu HL, Daxinger L, Pontes O, He X, Qian W, Lin H, Xie M, Lorkovic ZJ, Zhang S, Miki D, Zhan X, Pontier D, Lagrange T, Jin H, Matzke AJ, Matzke M, Pikaard CS, Zhu JK (2010) An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature 465:106–109
Gendrel AV, Lippman Z, Martienssen R, Colot V (2005) Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods 2:213–218
Haag JR, Pontes O, Pikaard CS (2009) Metal A and metal B sites of nuclear RNA polymerases Pol IV and Pol V are required for siRNA-dependent DNA methylation and gene silencing. PLoS ONE 4:e4110
Habu Y, Mathieu O, Tariq M, Probst AV, Smathajitt C, Zhu T, Paszkowski J (2006) Epigenetic regulation of transcription in intermediate heterochromatin. EMBO Rep 7:1279–1284
He XJ, Hsu YF, Zhu S, Liu HL, Pontes O, Zhu J, Cui X, Wang CS, Zhu JK (2009a) A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development. Genes Dev 23:2717–2722
He XJ, Hsu YF, Zhu S, Wierzbicki AT, Pontes O, Pikaard CS, Liu HL, Wang CS, Jin H, Zhu JK (2009b) An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell 137:498–508
He XJ, Hsu YF, Pontes O, Zhu J, Lu J, Bressan RA, Pikaard C, Wang CS, Zhu JK (2009c) NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev 23:318–330
Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobsen SE (2006) Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet 38:721–725
Herr AJ, Jensen MB, Dalmay T, Baulcombe DC (2005) RNA polymerase IV directs silencing of endogenous DNA. Science 308:118–120
Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008:420747
Ito H, Gaubert H, Bucher E, Mirouze M, Vaillant I, Paszkowski J (2011) An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472:115–119
Jackson JP, Johnson L, Jasencakova Z, Zhang X, PerezBurgos L, Singh PB, Cheng X, Schubert I, Jenuwein T, Jacobsen SE (2004) Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma 112:308–315
Johnson L, Cao X, Jacobsen S (2002) Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr Biol 12:1360–1367
Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE (2007) The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol 17:379–384
Johnson LM, Law JA, Khattar A, Henderson IR, Jacobsen SE (2008) SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet 4:e1000280
Jovtchev G, Borisova BE, Kuhlmann M, Fuchs J, Watanabe K, Schubert I, Mette MF (2011) Pairing of lacO tandem repeats in Arabidopsis thaliana nuclei requires the presence of hypermethylated, large arrays at two chromosomal positions, but does not depend on H3-lysine-9-dimethylation. Chromosoma 120:609–619
Kapoor A, Agarwal M, Andreucci A, Zheng X, Gong Z, Hasegawa PM, Bressan RA, Zhu JK (2005) Mutations in a conserved replication protein suppress transcriptional gene silencing in a DNA-methylation-independent manner in Arabidopsis. Curr Biol 15:1912–1918
Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11:204–220
Lawrence RJ, Pikaard CS (2004) Chromatin turn ons and turn offs of ribosomal RNA genes. Cell Cycle 3:880–883
Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE (2001) Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292:2077–2080
Mathieu O, Jasencakova Z, Vaillant I, Gendrel AV, Colot V, Schubert I, Tourmente S (2003) Changes in 5S rDNA chromatin organization and transcription during heterochromatin establishment in Arabidopsis. Plant Cell 15:2929–2939
Mathieu O, Probst AV, Paszkowski J (2005) Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J 24:2783–2791
Moghaddam AM, Roudier F, Seifert M, Berard C, Magniette ML, Ashtiyani RK, Houben A, Colot V, Mette MF (2011) Additive inheritance of histone modifications in Arabidopsis thaliana intra-specific hybrids. Plant J 67:691–700
Myouga F, Tsuchimoto S, Noma K, Ohtsubo H, Ohtsubo E (2001) Identification and structural analysis of SINE elements in the Arabidopsis thaliana genome. Genes Genet Syst 76:169–179
Naumann K, Fischer A, Hofmann I, Krauss V, Phalke S, Irmler K, Hause G, Aurich AC, Dorn R, Jenuwein T, Reuter G (2005) Pivotal role of AtSUVH2 in heterochromatic histone methylation and gene silencing in Arabidopsis. EMBO J 24:1418–1429
Pecinka A, Dinh HQ, Baubec T, Rosa M, Lettner N, Mittelsten SO (2010) Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 22:3118–3129
Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi MA, Lerbs-Mache S, Colot V, Lagrange T (2005) Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev 19:2030–2040
Rajakumara E, Law JA, Simanshu DK, Voigt P, Johnson LM, Reinberg D, Patel DJ, Jacobsen SE (2011) A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo. Genes Dev 25:137–152
Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593–599
Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53:247–259
Rowley MJ, Avrutsky MI, Sifuentes CJ, Pereira L, Wierzbicki AT (2011) Independent chromatin binding of ARGONAUTE4 and SPT5L/KTF1 mediates transcriptional gene silencing. PLoS Genet 7:e1002120
Slotkin RK, Vaughn M, Borges F, Tanurdzic M, Becker JD, Feijo JA, Martienssen RA (2009) Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136:461–472
Soppe WJ, Jasencakova Z, Houben A, Kakutani T, Meister A, Huang MS, Jacobsen SE, Schubert I, Fransz PF (2002) DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J 21:6549–6559
Stancheva I (2005) Caught in conspiracy: cooperation between DNA methylation and histone H3K9 methylation in the establishment and maintenance of heterochromatin. Biochem Cell Biol 83:385–395
Thorstensen T, Grini PE, Aalen RB (2011) SET domain proteins in plant development. Biochim Biophys Acta 1809:407–420
Wierzbicki AT, Haag JR, Pikaard CS (2008) Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135:635–648
Wierzbicki AT, Ream TS, Haag JR, Pikaard CS (2009) RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet 41:630–634
Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2:E104
Zilberman D, Cao X, Jacobsen SE (2003) ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299:716–719
Acknowledgments
We thank Beate Kamm, Christa Fricke and Inge Glaser for excellent technical assistance, Armin Meister and Rhonda Meyer for support with statistical data analysis and Ingo Schubert for critically reading the manuscript. We are grateful to Gunter Reuter for contribution of transgenic A. thaliana line containing the Pro35S-mycSUVH2 construct and critical reading of the manuscript. This work was supported by German research foundation (DFG) collaborative research centre (SFB) 648 “Molecular Mechanisms of Information Processing in Plants”.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Kuhlmann, M., Mette, M.F. Developmentally non-redundant SET domain proteins SUVH2 and SUVH9 are required for transcriptional gene silencing in Arabidopsis thaliana . Plant Mol Biol 79, 623–633 (2012). https://doi.org/10.1007/s11103-012-9934-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-012-9934-x