Abstract
The last steps of the Leu biosynthetic pathway and the Met chain elongation cycle for glucosinolate formation share identical reaction types suggesting a close evolutionary relationship of these pathways. Both pathways involve the condensation of acetyl-CoA and a 2-oxo acid, isomerization of the resulting 2-malate derivative to form a 3-malate derivative, the oxidation-decarboxylation of the 3-malate derivative to give an elongated 2-oxo acid, and transamination to generate the corresponding amino acid. We have now analyzed the genes encoding the isomerization reaction, the second step of this sequence, in Arabidopsis thaliana. One gene encodes the large subunit and three encode small subunits of this enzyme, referred to as isopropylmalate isomerase (IPMI) with respect to the Leu pathway. Metabolic profiling of large subunit mutants revealed accumulation of intermediates of both Leu biosynthesis and Met chain elongation, and an altered composition of aliphatic glucosinolates demonstrating the function of this gene in both pathways. In contrast, the small subunits appear to be specialized to either Leu biosynthesis or Met chain elongation. Green fluorescent protein tagging experiments confirms the import of one of the IPMI small subunits into the chloroplast, the localization of the Met chain elongation pathway in these organelles. These results suggest the presence of different heterodimeric IPMIs in Arabidopsis chloroplasts with distinct substrate specificities for Leu or glucosinolate metabolism determined by the nature of the different small subunit.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Leu, Val and Ile form a small group of amino acids distinguished by their branched aliphatic side chains. They are essential nutrients in human diets but can be de novo synthesized by bacteria, archaea, plants and fungi (Xing and Whitman 1991; Singh 1999; Kohlhaw 2003). In all species investigated the biosynthesis of these amino acids follows the same general pattern. Val and Ile are formed in two parallel pathways using the same enzymes while Leu formation branches off from 2-oxoisovalerate, the last intermediate of the Val biosynthetic pathway. However, in different organisms and kingdoms special features of branched-chain amino acid formation have been found involving the regulation and compartmentalization of the reaction cascades.
In plants all enzymatic activities required for the formation of branched-chain amino acids have been detected (Singh 1999; Binder et al. 2007), but only some of the required enzymes and genes have been characterized in detail. Analysis of the enzymes involved in plant amino acid metabolism has greatly benefited from complete genome sequences of model species which reveal candidate genes encoding proteins contributing to these pathways by their similarity to bacterial counterparts (The Arabidopsis Genome Initiative 2000). However, in the model species Arabidopsis thaliana (hereafter referred to as Arabidopsis), a clear assignment of branched-chain amino acid biosynthetic genes is more difficult than initially thought. This is particularly evident for Leu biosynthesis. The formation of this amino acid requires a three-step chain elongation, including a condensation of a 2-oxo acid with acetyl-CoA, an isomerization and an oxidation-decarboxylation, catalyzed by isopropylmalate synthase (IPMS), isopropylmalate isomerase (IPMI, also called a dehydratase) and isopropylmalate dehydrogenase (IPMDH), respectively (Fig. 1). Leu biosynthesis is then completed by a transamination step catalyzed by a branched-chain aminotransferase that could be also involved in the formation of the other branched-chain amino acids. In Arabidopsis and other Brassicaceae the same reaction cascade is also found in the Met chain elongation pathway (Fig. 1), the first part of the biosynthesis of aliphatic glucosinolates (Wittstock and Halkier 2002; Grubb and Abel 2006; Halkier and Gershenzon 2006; Binder et al. 2007). This similarity was recognized a long time ago (Strassman and Ceci 1963; Underhill et al. 1973) and the analysis of isopropylmalate synthases in Arabidopsis confirmed the strong relationship between the Leu biosynthesis and Met chain elongation pathways (Kroymann et al. 2001; Field et al. 2004; de Kraker et al. 2007; Textor et al. 2007). The two “true” isopropylmalate synthases (IPMS1 and 2) involved in the formation of Leu and the two methylthioalkylmalate synthases (MAM1 and 3) active in the chain elongation pathway of Met are encoded in the same gene family. While there seems to be a clear assignment of the IPMS proteins to Leu biosynthesis and the MAM proteins to Met chain elongation, this is not true for the aminotransferase step. The branched-chain aminotransferases (BCAT) 3 and 4 participate in Met chain elongation while BCAT3 is also active in branched-chain amino acid (BCAA) biosynthesis (Schuster et al. 2006; Knill et al. 2008).
Chemical structures of the intermediates of the later steps of Leu biosynthesis and Met chain elongation showing the parallel nature of the two pathways. The reaction types are the same, and the genes and enzymes are very similar. Up to six rounds of chain elongation are known to occur in Arabidopsis. Depicted are the first and second cycles. The elongated keto acid produced by each cycle (for example, 5-methylthio-2-oxopentanoate, 5MTOP, by the first round) can participate in a subsequent round of elongation, or can be transaminated to form an elongated Met derivative and then proceed to the later steps of glucosinolate biosynthesis
In contrast to the progress made in the characterization of IPMS and BCAT proteins, virtually nothing is known about the other two steps of the chain elongation sequence, isopropylmalate isomerase and isopropylmalate dehydrogenase (Singh 1999). In Arabidopsis there are two small gene families each encoding several proteins with sequence similarity to these previously characterized enzymes from other kingdoms (Schuster et al. 2006; Binder et al. 2007). It has been speculated that the different members of these families might function in Leu biosynthesis and/or Met chain elongation (Kroymann et al. 2001; Schuster et al. 2006; Binder et al. 2007). We have now analyzed genes encoding the large and small subunits of the isopropylmalate isomerase in Arabidopsis. Detailed metabolite profiling of several mutants revealed an involvement of the large subunit in both the biosynthesis of Leu and of chain-elongated Met derivatives. In contrast, there seems to be a specialization in the function of the small subunits. We also present experimental evidence that one of the small subunits involved in Met chain elongation is imported into chloroplasts providing further evidence that the complete Met chain elongation pathway is located in these organelles.
Materials and methods
Plant cultivation
Plants used for amino acid and glucosinolate analyses were cultivated as reported before (Knill et al. 2008).
Metabolite analysis
The contents of amino acids and glucosinolates were measured in approximately 3 week-old rosette leaves and seeds according to methods described previously (Roth 1971; Sarwar and Botting 1993; Brown et al. 2003; de Kraker et al. 2007; Knill et al. 2008). Between 5 and 15 replicates were analyzed for the individual lines. Plant material was obtained from a single cultivation per line. Details are given in Supplemental Table S1. Additionally, the flow-through fractions of the anion exchange chromatography performed in the course of glucosinolate analysis of leaf extracts of mutant and wild-type plants were compared by LC-MS using a Bruker Esquire 6,000 ion trap mass spectrometer (Bruker Daltonics, Bremen, Germany) operated in negative ion mode in the range m/z 50–1,000 (Skimmer voltage, −40 eV; capillary exit voltage, −104.1 eV; capillary voltage, 4,000 V; nebulizer pressure, 40 psi; drying gas, 11 l min−1; gas temperature, 330°C) coupled to an Agilent 1100 series HPLC (Agilent Technologies, Waldbronn, Germany). Elution was accomplished using a Nucleodur Sphinx RP column (250 × 4.6 mm, 5 μm; Macherey-Nagel, Düren, Germany). Mobile phases were 0.2% formic acid (v:v) (A) and acetonitrile (B), starting with 100% A for 5 min, followed by a gradient to 75% B in 25 min.
The subtraction of the mass spectrometer total ion chromatogram of wild-type plants from that of the mutants (using the software package Metabolite Detect 1.1, Bruker Daltonics, Bremen, Germany) identified two additional peaks in ipmi lsu1-3 extracts not found in wild-type extracts corresponding to molecular masses of 176 (compound 1) and 238 (compound 2). The former was unambiguously identified as 2-isopropylmalate (2-IPM) by identical retention times with an authentic 2-IPM standard (Aldrich) in HPLC and identical fragmentation patterns in MS2 experiments in the negative ionization mode as described above.
Spectroscopic identification of compound 2, 2-(3′-methylsulfinyl)propylmalate
2-(3′-Methylsulfinyl)propylmalate (compound 2) was isolated from aqueous leaf extracts from ipmi lsu1-3 plants by preparative HPLC and analyzed by mass spectrometry as described above, both positive and negative ionization modes were employed. High resolution electrospray ionization (ESI) mass spectra were recorded on a Bruker microTOF mass spectrometer (Bruker Daltonics, Bremen, Germany). NMR spectra were recorded on a Bruker AV 500 spectrometer (Bruker Biospin, Rheinstetten, Germany) equipped with a CryoPlatform. 1H NMR, 1H–1H COSY, HSQC and HMBC spectra were recorded using a 5 mm TCI CryoProbe. The operating frequencies were 500.130 MHz for acquisition of 1H and 125.758 MHz for 13C spectra. Samples were measured at 293 K in methanol-d 4. Chemical shifts are given in ppm, referenced to the remaining solvent signals (3.31 ppm for 1H signals and 49.05 ppm for 13C signals).
2-(3′-Methylsulfinyl)propylmalate (2), 1H and 13C NMR (see Supplemental Table S2) Low resolution ESI-MS: m/z 238.9 [M + H]+ and m/z 237.0 [M−H]− MS2 experiments with LRESIMS resulted in following fragments: fragments of m/z 238.9 [M + H]+: 193.0 [M−CO2H2]+ 31.8%, 174.9 [M–CH3–SHO]+ 38.3%, 129.0 [M–CO2H2–CH3–SHO]+ 6.7%; fragments of m/z 237.0 [M−H]−: 221.8 [M−CH3]− 7.5%, 176.9 [M–CH3–CO2H]− 28.9%, 172.9 [M–CH3–SHO]− 33.9%. The molecular formula of compound 2 was determined to be C8H14O6S (calculated monoisotopic mass 238.0511) from the [M−H]− ion peak at m/z 237.0427 in high resolution ESI-MS. NMR spectroscopy (1H, COSY, HSQC, HMBC) revealed 8 carbon signals of which two low field resonances at 180.3 and 175.6 ppm indicated the carboxylic functions of the substituted malate, followed by another quaternary carbon resonance at 75.5 ppm (position 2 of the malate). In the proton spectrum only the signal at 2.70 ppm is well resolved (doublet of doublets, H-3a/b). All other proton resonances are higher order multiplets. The connectivity was deduced from prominent resonances in the HMBC spectrum for H-3a/b and the S-methyl singlet, as well as by COSY cross peaks (Supplemental Table S2).
Nucleic acid analysis
Total DNA and RNA, respectively, were isolated as described previously (Forner et al. 2007). PCR, RT-PCR and Northern blot hybridization were done following established protocols (Sambrook and Russel 2001). Oligonucleotide sequences are available on request. Real-Time quantitative RT-PCR was performed using Transcriptor High Fidelity cDNA Synthesis and Light-Cycler 480 SYBR Green I Master Kits on a Light-Cycler 480 Real-Time PCR System according to instruction given by the manufacturer (Roche). Data of three samples were evaluated using the Light-Cycler 480 software (1.5). Relative transcript levels were measured with respect to validated reference genes encoding ubiquitin conjugating enzyme 9 (UBC9, At4g27960) and protein phosphatase 2A subunit 3 (PP2A, At1g13320) with established primer pairs (Czechowski et al. 2005). Oligonucleotides for the IPMI LSU1 expression analysis were selected using the primer design tool at the NCBI home page (http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC = BlastHomeAd).
Miscellaneous methods
Standard methods of molecular biology were done as described (Sambrook and Russel 2001). GFP tagging experiments were performed following established procedures (Koop et al. 1996; Däschner et al. 2001). DNA, cDNA and proteins sequences were investigated using the blast tools and Clustal W.
Results
Identification of genes for large and small subunits of isopropylmalate isomerase (IPMI)
Isopropylmalate isomerase (IPMI) in bacteria is present as a dimer, whose subunits are distinct and vary almost twofold in size (Gruer et al. 1997; Kohlhaw 2003). The Arabidopsis IPMI has a similar structure. The genome contains a single gene for the large subunit (At4g13430, IPMI LSU1) and three genes for the small subunits of this enzyme (At2g43090 (IPMI SSU1), At2g43100 (IPMI SSU2), and At3g58990 (IPMI SSU3))(Schuster et al. 2006; Binder et al. 2007) whose sequences align closely with the respective sequences from E. coli (leuC and leuD) and an isopropylmalate isomerase of the single subunit type found in fungi (Gruer et al. 1997; Kohlhaw 2003) (Supplemental Fig. S1). The proteins from Arabidopsis have 34% identity at the amino acid level to their counterparts from E. coli. The Arabidopsis large subunit also aligns well to the N-terminal part of the single subunit type of IPMI from Candida albicans (LEU1), and the small subunit aligns well with the C-terminal part. The alignments of both the Arabidopsis large and small subunits to their respective microorganism counterparts also revealed N-terminal extensions which might function as plastid targeting signals.
Characterization of mutants for the large subunit of the IPMI
To obtain information about the function of the large subunit of the isopropylmalate isomerase in vivo, we investigated three independent T-DNA insertion mutants obtained from the SALK, SAIL and GABI-Kat collections (Sessions et al. 2002; Alonso et al. 2003; Rosso et al. 2003) Genotyping of respective plants and sequencing of the T-DNA flanking sequences identified the T-DNAs within the 5′ UTR of the gene either about 50 (SALK 029510 and SAIL 369G04) or eleven nucleotides upstream of the translation start codon (GABI Kat 452D03, Fig. 2 and Supplemental Fig. S2). These T-DNA alleles were designated ipmi lsu1-1 (SALK 029510), ipmi lsu1-2 (SAIL 369G04) and ipmi lsu1-3 (GABI Kat 452D03).
Characterization of three independent IPMI LSU1 mutants: The gene encoding the large subunit of isopropylmalate isomerase1 (IPMI LSU1, At4g13430) contains 15 exons (black boxes). The first three exons (E1 to E3) are enlarged and the approximate localizations of the T-DNA insertions of three independent lines and of the oligonucleotides (vertical arrows) used for genotyping are given. Further details are given in Supplemental Fig. S2
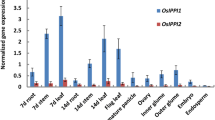
Transcript analysis of homozygous ipmi lsu1-1, 1-2 and 1-3 plants by RT-PCRs covering the complete reading frame detected correctly spliced IPMI LSU1 mRNA suggesting that the T-DNA insertions in the 5′ region do not yield knockouts (data not shown). But the fact that the T-DNA insertions in these lines are located just downstream of the transcription initiation site imply that their presence might influence transcription of the IPMI LSU1 gene in these mutants. As expected, quantitative Real-Time RT-PCR revealed that the steady state levels of the IPMI LSU1 transcripts in these mutants are substantially reduced in seedlings and 3 week-old rosette leaves (Fig. 3). In the ipmi lsu1-2 mutant mRNA levels are reduced to 65 and 50% of the wild-type standard in seedlings and rosette leaves, respectively, while IPMI LSU1 transcript levels are reduced to 40 and 27% of wild type in these tissues of ipmi lsu1-1 plants. Strongest reduction is seen in ipmi lsu1-3 seedlings (21% of wild type), while the IPMI LSU1 mRNA level in ipmi lsu1-3 reaches about 29% of the wild-type level in leaves. Thus all insertions cause a reduced IPMI LSU1 steady-state transcript level, which is most severe in the ipmi lsu1-3 mutant. Further cDNA analysis revealed that transcription of IPMI LSU1 gene in ipmi lsu1-1 and 1-3 plants starts within the T-DNA insertion (data not shown). While the insertion in ipmi lsu1-1 has no effect on the IPMI LSU1 reading frame, the insertion 11 nucleotides upstream of the ATG in the ipmi lsu1-3 mutant introduces another in frame ATG and thus extends the reading frame at the N-terminus (Supplemental Fig S2B). Moreover the insertion in this plant line promotes a severe delay in development, which is not observed for the ipmi lsu1-1 and -2 mutants (Supplemental Fig. S3, data not shown).
The T-DNA insertions in the 5′ UTR of the IPMI LSU1 gene change transcript levels in different tissues. Real-Time quantitative RT-PCR was performed three times on total RNA obtained from seedlings, flowers and rosette leaves of 3 week-old ipmi lsu1-1, ipmi lsu1-2 ipmi lsu1-3 and wild-type plants. Transcript levels in these mutants (ipmi lsu1-1: dark gray column; ipmi lsu1-2: light grey column, ipmi lsu1-3: white column) are given relative to the mRNA levels measured in wild type (black column)
Amino acid profiles of large subunit mutants
To investigate the function of the IPMI LSU1 gene in vivo, the levels of free amino acids were determined in 3 week-old plants and seeds of the three homozygous ipmi lsu1 mutants and wild-type controls. In leaves, ipmi lsu1-2 plants exhibit the weakest chemical phenotype with only two significant changes in comparison to wild type (Supplemental Table S3). In contrast, many more changes were seen between the ipmi lsu1-1 and 1-3 plants and wild type. Most striking is the accumulation of S-methylmethionine (SMM) in rosette leaves of both mutants (0.19 ± 0 and 0.26 ± 0.05 nmol/mg dry weight, respectively). This is accompanied by twofold increased levels of Met (0.41 ± 0.07 in ipmi lsu1-1 vs. 0.19 ± 0 in wild type and 0.39 ± 0.07 in ipmi lsu1-3 vs. 0.18 ± 0.06 nmol/mg dry weight in wild type). Of the branched-chain amino acids, only Val is significantly elevated in the ipmi lsu1-3 mutant (1.27 ± 0.08 in ipmi lsu1-3 vs. 0.84 ± 0.14 nmol/mg dry weight in wild type), whereas no alterations were detected in Leu as well as Ile levels. Apart from that other amino acids show almost exclusively minor alterations.
In seeds, many significant but mostly moderate changes were found when comparing among the mutants and wild type (Supplemental Table S4). These alterations do not reveal any clear pattern, except that SMM was found in the ipmi lsu1-1 and 1-3 mutants, while this transport or storage form of Met is barely detectable in ipmi lsu1-2, and is not observed in wild-type seeds (ipmi lsu1-1: 0.42 ± 0.31; ipmi lsu1-2: 0.04 ± 0.03; ipmi lsu1-3: 1.33 ± 0.75 nmol/mg seeds).
The observed variations in the amino acid and glucosinolate contents (Tables 1, 2) among the different wild-type lines are likely due to the fact that not all plants were grown at the same time. Even very minor variations in the experimental growth conditions can lead to alterations in metabolites, such as glucosinolates or amino acids. However, all mutants and their corresponding wild-type lines were always grown together and analyzed in parallel, which allows unambiguous conclusions about the effect of the gene knockout.
Intermediates of Leu biosynthesis and Met elongation accumulate in IPMI large subunit mutants
Although the amino acid profiles did not indicate a function of IPMI LSU1 in Leu biosynthesis, we searched for further evidence on this point by LC-MS analysis of methanolic extracts obtained during glucosinolate isolation. Comparison of the ipmi lsu1-3 chromatogram with that obtained from wild-type leaf extracts revealed two extra peaks in the mutant corresponding to molecular masses of 176 (compound 1) and 238 (compound 2). The metabolite with a molecular mass of 176 was identified to be the enzyme substrate, 2-isopropylmalate (2-IPM, Fig. 4) by similarity of retention time and MS fragmentation to those of authentic 2-IPM. A quantitative analysis of all three IPMI LSU1 mutants revealed an average 2-IPM content of 0.42 and 0.02 mg/g dry weight in leaves of ipmi lsu1-3 and ipmi lsu1-1 mutants, respectively, while this metabolite was undetectable in the ipmi lsu1-2 plants.
Compound 2 was identified by spectroscopic methods as 2-(3′-methylsulfinyl)propylmalate (Fig. 4, for details see Material and Methods), an oxidation product of 2-(3′-methylthio)propylmalate, an intermediate in the second round of Met chain elongation (Fig. 1). This compound is formed by condensation of 5-methylthio-2-oxopentanoate (5MTOP) and acetyl-CoA, catalyzed by a methylthioalkylmalate synthase (MAM1 or MAM3). In contrast to 2-IPM, this metabolite accumulates in all three mutants, while it is not seen in wild type. Relative levels of 2-(3′-methylsulfinyl)propylmalate are 10/1/14 in ipmi lsu1-1, 1-2 and 1-3 plants.
In summary the accumulation of 2-IPM and 2-(3′-methylsulfinyl)propylmalate in the ipmi lsu1 mutants indicates a function of IPMI LSU1 in both the biosynthesis of Leu and in the Met chain elongation pathway of glucosinolate formation.
Glucosinolate composition is shifted towards shorter chain species in large subunit mutants
To gain more information on the role of IPMI LSU1 in glucosinolate biosynthesis, the profiles of aliphatic glucosinolates in ipmi lsu1 mutants and wild-type lines were compared in leaves of 3 week-old plants and in seeds. The mutants exhibited a strong trend towards increases in short-chain glucosinolates accompanied by reductions in long-chain aliphatic glucosinolates (Tables 1, 2). In both leaves and seeds, all C3 glucosinolates were amply increased, while all C7 and C8 glucosinolate species are strongly reduced. In seeds also 5-methylthiopentylglucosinolate (5MTP) is reduced, while the decrease of 5-methylsulfinylpentylglucosinolate (5MSOP) has only been measured in ipmi lsu1-1. This redistribution of glucosinolate chain length is particularly strong in the ipmi lsu1-3 mutant, where all C5, C7 and C8 glucosinolates are reduced to undetectable amounts in both seeds and leaves. The C4 glucosinolates can be either increased or decreased depending on the compound and mutant.
In summary, these metabolite profiles, along with the accumulation of SMM, constitute strong evidence for the participation of the IPMI LSU1 gene in the biosynthesis of aliphatic glucosinolates.
Characterization of mutants for the small subunit of IPMI
In our study of the IPMI SSU genes, we investigated two mutants for IPMI SSU1 (SALK 115589, here called ipmi ssu1-1, and SALK 111666, here ipmi ssu1-2), and one mutant each for IPMI SSU2 (SALK 548320, ipmi ssu2-1) and IPMI SSU3 (FLAG 149F10, ipmi ssu3-1). For IPMI SSU1, no plants homozygous for the T-DNA allele could be obtained in either analyzed line. When siliques from heterozygous plants were inspected, they exhibited abnormalities in seed maturation. In line SALK 115589 seed development seems to be aborted at a very early stage, while seed maturation is interrupted at a later phase in SALK 111666 (Supplemental Fig S4). Since in both independent lines the number of mature seeds is substantially reduced and no homozygous mutants could be obtained, an intact IPMI SSU1 gene seems to be crucial for the development of mature viable seeds.
In contrast, homozygous ipmi ssu2-1 and 3-1 mutants could be established and the presence of the T-DNA insertion was confirmed near the center and the beginning of the reading frames, respectively (Fig. 5a, c). Northern analysis of ipmi ssu2-1 (Fig. 5b) and RT-PCR assays of ipmi ssu3-1 plants (Fig. 5d) revealed the absence of mature mRNAs in the homozygous plants and the knockout of these genes by the T-DNAs. Both lines, which did not show any macroscopic phenotype, were then cultivated for metabolite analyses.
Characterization of ipmi ssu2-1 and ipmi ssu3-1 mutants. T-DNA insertions were confirmed in line SALK 048320 near the center of the IPMI SSU2 gene a and in line FLAG 149F10 within the ATG of the IPMI SSU3 c reading frame (black boxes). Oligonucleotides used for genotyping (not shown) and RT-PCR are given as vertical arrows. Left border sequences (LB) were detected at both extremities of the insertion in SALK 048320. b A northern blot hybridization detected a 0.8 kb mRNA in wild type (wt) and heterozygous plants (ht, reduced level), while the lack of this transcript demonstrates the knockout of the IPMI SSU2 gene in homozygous (lane hm) ipmi ssu2-1 plants. Ethidium bromide staining of the rRNAs shows the quality of the RNA samples and the equal loading of the gel. d RT-PCR using primers for IPMI SSU3 detected product of the expected size (~300 bp) in wild-type cDNA (lane wt), but not in cDNA from homozygous ipmi ssu3-1 plants (lane ko). A control RT-PCR (UBQ10) was performed as described in legend of Fig. 2
The free amino acid content of the leaves and seeds of ipmi ssu2-1 plants was not substantially different from that of the wild type (Supplemental Tables S5 and S6). The statistically significant changes were only 1.1- to 1.3-fold. No significant alterations were seen for Leu, while Ile was slightly decreased in leaves and very moderately elevated in seeds, and Val was found to be 1.2-fold enriched in seeds. Met levels, often changed in mutants compromised in other steps of the Met chain elongation pathway, were unaffected. In addition, SMM, which usually accumulates in Met chain elongation mutants, was not detectable in the ipmi ssu2-1 mutant.
The profile of the methionine-derived glucosinolates in leaves of ipmi ssu2-1 also did not show substantial variations from that of wild-type plants (Supplemental Table S7). Only an increase in 7MSOH and decreases in 8MSOO and I3M are observed. In contrast, seeds exhibited clear elevations of total glucosinolate content as well as total methionine-derived glucosinolate content (between 1.3- and 2.4-fold, Supplemental Table S8). Apart from 4-benzoyloxybutylglucosinolate (4BZO, 92%) all C3 and C4 glucosinolates are elevated (1.5- to 1.7-fold) in the mutant. In terms of absolute values, the most prominent increase was observed for 4MTB, the main glucosinolate species in seeds of Arabidopsis accession Columbia (Col). The increase of these short-chain glucosinolates is by trend similar to the elevated levels of these glucosinolate species in the ipmi lsu1 mutants. In contrast to the latter long-chain glucosinolates are either increased (C7) or decreased (C8).
In leaves and seeds of the ipmi ssu3-1 mutant (which has a Wassilewskija genetic background) there were also few significant changes in amino acid content (Supplemental Table S9 and S10). In leaves, reductions were found for Gln (85% of wild-type level) and Gly (52%), which help account for the reduction of the total amount of free amino acids (88% of wild-type level). In seeds, moderate increases were measured for Asn, Ile, Leu and Ser (between 1.2- and 1.4-fold). As in ipmi ssu2-1, Met levels were unchanged in ipmi ssu3-1 and SMM could not be detected. Profiling of glucosinolates in both leaves and seeds also did not reveal any substantial changes from wild-type levels (Supplemental Tables S11 and S12).
The small subunit of IPMI is imported into chloroplasts
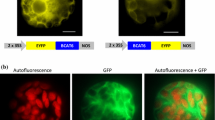
Both the biosynthesis of Leu and the Met chain elongation pathway are known to occur in chloroplasts (Falk et al. 2004; Binder et al. 2007; Textor et al. 2007; Knill et al. 2008) suggesting that the IPMI subunits must be imported into these organelles. Consistent with this suggestion, all four IPMI subunits of Arabidopsis are predicted to have chloroplast targeting sequences by amino acid sequence alignments and an in silico analysis of subcellular sorting by two prediction servers (Predotar: http://urgi.versailles.inra.fr/predotar/predotar and TargetP: http://www.cbs.dtu.dk/services/TargetP) (data not shown). To experimentally verify these predictions, the full length reading frame of one of the small subunit genes (IPMI SSU2, At2g43100) was fused in frame to the 5′ end of the gene encoding the green fluorescent protein (GFP) in the vector psmGFP4 (Davis and Vierstra 1998). This construct was transiently expressed in tobacco protoplasts and stably transformed into Arabidopsis plants. Transformed protoplasts from tobacco and protoplasts released from leaves of transgenic Arabidopsis plants were then inspected by fluorescence microscopy. The patterns of GFP fluorescence and autofluorescence were identical demonstrating an import of the IPMI SSU2:GFP fusion protein into chloroplasts (Fig. 6). This indicates that the native IPMI SSU2 protein is also imported into chloroplasts in vivo.
Subcellular localization of the small subunit of isopropylmalate isomerase encoded by At2g43100. The full length reading frame of IPMI SSU2 was fused in frame 5′ to the GFP gene (bottom part). The fusion protein was transiently expressed in tobacco protoplasts (N.t., middle part) and in stable transgenic Arabidopsis plants (A.t., top part). Bars = 20 μm. Two filter sets were used to detect the fluorescence of GFP and chlorophyll or of GFP alone
Discussion
The large IPMI subunit is involved in both Leu and glucosinolate metabolism, while the small subunits appear to be specific for each pathway
Arabidopsis possesses genes whose sequences align closely with the isopropylmalate isomerase genes of E. coli (leuC and leuD, Supplemental Fig. S1), indicating that this leucine biosynthetic enzyme probably has the same heterodimeric structure in both of these distantly related species. The pattern is also likely to extend throughout the plant kingdom given the similarity of Arabidopsis and rice sequences to each other. To confirm the biochemical function of the Arabidopsis isopropylmalate isomerase, lines with insertional mutations in the corresponding genes were characterized and chemically analyzed.
Mutant lines with lower transcript levels of the single gene encoding the large subunit of isopropylmalate isomerase (IPMI LSU1) demonstrate that this protein plays an important role in the biosynthesis of Leu (Figs. 2, 3, 7). The IPMI substrate, 2-IPM, accumulates when transcript of IPMI LSU1 is reduced sufficiently. Although the actual level of Leu remains unchanged in comparison to wild type, there is a significant increase in Val (Supplemental Table 3), most likely due to a reduced flux from the Val pathway into Leu biosynthesis.
Leu biosynthesis and Met chain elongation pathways. Met chain elongation pathway is shaded in middle grey, the Leu biosynthesis pathway in dark grey and the pathways leading to Val and Ile are highlighted in light grey. Enzymes are given in boxes and are abbreviated as follows: AHAS: acetohydroxyacid synthase; BCAT: branched-chain aminotransferase; DHAD: dihydroxyacid dehydratase; IPMI LSU: isopropylmalate isomerase large subunit; IPMI SSU: isopropylmalate isomerase small subunit; IPMDH: isopropylmalate dehydrogenase; IPMS: isopropylmalate synthase; KARI: ketolacid reductoisomerase; MAM: methylthioalkylmalate synthase. MAM1/2* indicates that either MAM1 (in Col-0) or MAM2 (in Landsberg erecta) catalyze condensation reactions leading to the preferential accumulation of C4 (in Col-0) and C3 (in Landsberg erecta) glucosinolates. The IPMI is highlighted by black boxes. Abbreviations for metabolites are: Pyr: pyruvate; 2OB: 2-oxobutyrate; 2AL: 2-acetolactate; 2A2HB: 2-aceto-2-hydroxybutyrate; 2,3DH3 MB: 2,3-dihydroxy-3-methylbutyrate; 2,3DH3 MP: 2,3-dihydroxy-3-methylpentanoate; 3 MOB: 3-methyl-2-oxobutanoate; 3 MOP: 3-methyl-2-oxopentanoate; 4MOP: 4-methyl-2-oxopentanoate; 2-IPM: 2-isopropylmalate; 3-IPM: 3-isopropylmalate; 4MTOB: 4-methylthio-2-oxobutanoate; MTOA: 4-methylthio-2-oxoacid
The large subunit of Arabidopsis IPMI also participates in the Met chain elongation reactions of aliphatic glucosinolate formation as indicated by the accumulation of SMM in mutant lines, a trait observed for mutants in other genes encoding enzymes of Met chain elongation (Fig. 7) (Schuster et al. 2006; Knill et al. 2008). This function is further supported by a shift in glucosinolate composition to shorter chain-length species (Tables 1, 2) and the accumulation of 2-(3′-methylsulfinyl)propylmalate. This metabolite is an oxidation product (enzymatic or non-enzymatic) of the IPMI substrate for the second round of chain elongation, 2-(3-(methylthio)propylmalate. Moreover, the IPMI LSU1 gene is known to be under the control of MYB28, a regulator of many genes involved in biosynthesis of Met-derived aliphatic glucosinolates (Gigolashvili et al. 2007; Hirai et al. 2007; Sonderby et al. 2007).
The ability of the large subunit of IPMI to participate in catalyzing reactions of substrates from both Leu biosynthesis and the Met chain elongation cycle may be accomplished by its interaction with different IPMI small subunits of which three are encoded in the Arabidopsis genome. In bacteria, the IPMI small subunits are involved in substrate binding (Gruer et al. 1997). The lethal phenotype of homozygous ipmi ssu1-1 plants suggests a unique function of this protein, most likely in Leu biosynthesis which cannot be compensated by either of the other two small subunits of IPMI (IPMI SSU2 and SSU3, Fig. 7). Instead these latter two proteins seem to be functionally redundant as indicated by the relatively weak phenotype of the respective mutants. In the seeds of the ipmi ssu2-1 mutant short-chain glucosinolates are increased similarly to what has been observed in the ipmi lsu1 mutants. This shift towards short-chain products seems to be a general phenomenon when the isomerase step is impaired. The very weak phenotype of the ipmi ssu3-1 might be due to this mutant being in the Ws-0 background, which per se contains predominantly short-chain glucosinolates (Supplemental Table S12).
In IPMI mutants in the Col background (ipmi ssu2-1, ipmi lsu1-1 and ipmi lsu1-2) the shift towards the short-chain glucosinolates is accompanied by an increase in total aliphatic glucosinolates (Table 2, Supplemental Table S8), which might be explained by different substrate specificities of other enzymes in the glucosinolate biosynthesis pathway. In contrast to ipmi lsu1 mutants, the increase in total aliphatic glucosinolates is particularly strong in the ipmi ssu2-1 seeds, since there is no reduction of the long-chain products (Supplemental Table S8).
Nevertheless the alterations in the glucosinolate composition observed in these mutants suggest that IPMI SSU2 and IPMI SSU3 function in Met chain elongation (Fig. 7), which is further supported by the strong co-expression of the corresponding genes with MYB28, MAM1 and BCAT4 (Hirai et al. 2007; Sonderby et al. 2007). In contrast, the other small subunit, IPMI SSU1, is not co-expressed with these genes.
The large subunit mutants have distinct phenotypes
The three ipmi lsu1 mutants studied all exhibit a shift towards shorter chain-length aliphatic glucosinolates. However, there are differences between the individual mutants in other traits. For example, the ipmi lsu1-2 plants showed the smallest chemical differences from wild-type plants. Changes in amino acid and glucosinolate levels were moderate, SMM and 2-IPM did not accumulate to detectable levels, and 2-(3′-methylsulfinyl)propylmalate content was the lowest among the three different ipmi lsu1 mutants (Tables 1, 2, Supplemental Tables 3 and 4). Accordingly, the IPMI LSU transcript level in this line showed the lowest reduction from wild type of the three mutants (Fig. 4).
A stronger chemical phenotype was observed in the ipmi lsu1-1 mutant, with SMM, 2-IPM and 2-(3′-methylsulfinyl)propylmalate accumulating to substantial amounts.
The strongest effects on both glucosinolate and amino acid composition as well as on the accumulation of 2-IPM and 2-(3′-methylsulfinyl)propylmalate were observed in ipmi lsu1-3 plants. However, the IPMI LSU1 transcript level in leaves of ipmi lsu1-3 plants was comparable to that in the ipmi lsu1-1 line. Thus the alterations in ipmi lsu1-3 cannot be solely explained by reduced transcript levels. As shown in Supplemental Fig S2B, the T-DNA insertion in ipmi lsu1-3 extends the IPMI LSU1 reading frame at the N-terminus. This lowers the calculated probability for an import into chloroplasts suggesting that the import of this protein into chloroplasts in vivo might be reduced in this mutant. In addition, the insertion of the T-DNA only 11 bp upstream of the ATG might also cause changes in spatio-temporal transcription of this gene or alter translation efficiency. These changes might be responsible for exacerbating the effect of the reduced IPMI LSU1 transcript. Plant growth could also be impaired by an expanded triplet repeat in the third intron of the IPMI LSU1 gene. This natural genetic defect, called ILL1 for irregularly impaired leaves in Arabidopsis accession Bur-0 results in a 50% reduction of the steady state mRNA level of this gene under certain growth conditions (Sureshkumar et al. 2009). Like the allele in Bur-0, ipmi lsu1-3 is also not a complete knockout allele. Since there is only a single gene encoding the large subunit of the potentially heterodimeric IPMI, the remaining IPMI LSU1 transcript is sufficient for some biosynthesis of Met-derived aliphatic glucosinolates and maintaining Leu levels comparable to wild-type plants. Leu levels are unchanged in all mutants despite the other metabolite alterations observed, indicating that amino acid levels are extraordinarily well-buffered against fluctuations.
The Met chain elongation cycle is located in the plastids
The biosynthesis of Met-derived aliphatic glucosinolates is distributed over several subcellular compartments. While the later phases such as core structure formation and side chain modifications proceed in the cytosol-endoplasmatic reticulum (Halkier and Gershenzon 2006), the compartmentalization of the Met chain elongation pathway appears to be more complex. The initial transamination reaction converting Met into 4-methylthio-2-oxobutanoate (4MTOB) occurs in the cytosol (Schuster et al. 2006). In contrast, most of the subsequent steps were suggested to be located in the plastids (Falk et al. 2004; Schuster et al. 2006) and there is now increasing evidence that this is indeed the case. For instance, the MAM3 enzyme catalyzing the condensation reaction yielding methylthioalkylmalate derivatives has been found to be located in plastids as is BCAT3, an aminotransferase required for the formation of short-chain aliphatic glucosinolates (Diebold et al. 2002; Textor et al. 2007; Knill et al. 2008). Here we provide evidence that the reaction following condensation, isopropylmalate isomerization, also occurs in plastids based on the import of IPMI SSU2 into chloroplasts where it can form a heterodimer with IPMI LSU1 (Fig. 6). In Arabidopsis the latter protein has also been detected in the stroma fraction of chloroplasts by a proteomic study, which also reported BCAT3, MAM1 and all three potential isopropylmalate dehydrogenases in protein fractions from plastids (Zybailov et al. 2008). Thus most of the proteins required for Met chain elongation are located in the chloroplast as are all enzymes of Leu biosynthesis.
The recruitment of Leu biosynthetic enzymes for the Met chain elongation pathway: dual roles vs. specificity
Leucine biosynthesis and the Met chain elongation pathway of glucosinolate biosynthesis share a very similar series of reactions (Figs. 1, 7). It has been previously suggested that Met chain elongation was evolutionarily derived from leucine formation, and recent investigations of the genes involved have shed light on how the recruitment of enzymes took place.
The initial step in chain elongation, the deamination of Met to 4MTOB is catalyzed by BCAT4 (Figs. 1, 7), an enzyme recruited from the aminotransferases of branched chain amino acid metabolism (Schuster et al. 2006). This protein has a clear preference for Met and Homo-Met (and their corresponding 2-oxo acids) over substrates of branched chain pathways. BCAT4 still shows in vitro activity with Leu, probably evidence of its evolutionary origin, but it is located in the cytosol, physically separated from the other enzymes of the Leu pathway (Knill et al. 2008; Schuster et al. 2006).
The next step in chain elongation is the condensation of 4MTOB with acetyl-CoA to produce a 2-(methylthio)alkylmalate derivative (Figs. 1, 7). In Arabidopsis Col-0, this reaction is catalyzed by two proteins, MAM1 and MAM3 that are specific for Met chain elongation and have no role in Leu biosynthesis. Similarly, the condensation reaction in Leu formation (reaction of 3 MOB with acetyl-CoA to produce 2-IPM) is catalyzed by two proteins, IPMS1 and IPMS2, that are specific for Leu biosynthesis and have no role in glucosinolate metabolism (de Kraker et al. 2007; Field et al. 2004; Textor et al. 2007).
In both Leu formation and Met chain elongation, the 2-alkylmalate condensation product is next isomerized to form a 3-alkylmalate derivative. These steps are catalyzed in part by the same enzyme, a departure from the pathway-specific enzymes of the previous two steps (Fig. 7). The Arabidopsis IPMI is most likely a heterodimer based on the sequence similarity to the heterodimeric IPMI proteins from bacteria, archaea and other plants (although this has not yet been proven by direct analysis of the proteins). Here we have demonstrated that the large subunit participates in both Leu and glucosinolate metabolism. IPMIs belong to the aconitase protein superfamily, whose members are characterized by four structural domains. In classical aconitases and IPMIs from fungi, all four domains are present in a single protein sequence (Gruer et al. 1997). However, in IPMIs of bacteria, archaea and plants domains 1–3 are found in a large subunit, while domain 4 is found in a small subunit. The overall structure of the heterodimeric IPMIs is very similar to that of the single subunit IPMIs and the aconitases. The crystal structure of the porcine mitochondrial aconitase revealed that domain 4 contributes to both substrate recognition and catalysis (Gruer et al. 1997). The crystal structure of a small subunit of IPMI from the archaea Pyrococcus horikoshii also allows the conclusion that the small subunit has a role in substrate recognition (Yasutake et al. 2004). Thus it is likely that the plant small subunit of IPMI influences substrate binding. Our demonstration that the Arabidopsis genome encodes three small subunits of IPMI, one of which seems specialized for Leu formation and the other two for Met chain elongation, supports the hypothesis that the small subunits determine the substrate specificities of the heterodimeric isopropylmalate isomerases in plants. Thus, the resulting IPMI heterodimer is pathway-specific even if the large subunit per se is not. These various IPMI heterodimers may then be differently distributed over different tissue types as a result of tissue-specific expression patterns of the small subunits.
As for the two remaining steps of Leu biosynthesis and Met chain elongation, the dehydrogenase and the final aminotransferase, more research is needed on the relevant genes and enzymes to make functional and evolutionary inferences. It will also be interesting to investigate how the subcellular and cellular localization of the two pathways and other regulatory aspects have been modified in evolutionary recruitment.
References
Alonso JM, Stepanova AN, Leisse TJ et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301:653–657
Binder S, Knill T, Schuster J (2007) Branched-chain amino acid metabolism in higher plants. Physiol Plant 129:68–78
Brown PD, Tokuhisa JG, Reichelt M, Gershenzon J (2003) Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62:471–481
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17
Däschner K, Couee I, Binder S (2001) The mitochondrial isovaleryl-coenzyme a dehydrogenase of Arabidopsis oxidizes intermediates of leucine and valine catabolism. Plant Physiol 126:601–612
Davis SJ, Vierstra RD (1998) Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol 36:521–528
de Kraker J-W, Luck K, Textor S, Tokuhisa JG, Gershenzon J (2007) Two Arabidopsis genes (IPMS1 and IPMS2) encode isopropylmalate synthase, the branchpoint step in the biosynthesis of leucine. Plant Physiol 143:970–986
Diebold R, Schuster J, Däschner K, Binder S (2002) The branched-chain amino acid transaminase gene family in Arabidopsis encodes plastid and mitochondrial proteins. Plant Physiol 129:540–550
Falk KL, Vogel C, Textor S, Bartram S, Hick A, Pickett JA, Gershenzon J (2004) Glucosinolate biosynthesis: demonstration and characterization of the condensing enzyme of the chain elongation cycle in Eruca sativa. Phytochemistry 65:1073–1084
Field B, Cardon G, Traka M, Botterman J, Vancanneyt G, Mithen R (2004) Glucosinolate and amino acid biosynthesis in Arabidopsis. Plant Physiol 135:828–839
Forner J, Weber B, Thuss S, Wildum S, Binder S (2007) Mapping of mitochondrial mRNA termini in Arabidopsis thaliana: t-elements contribute to 5′ and 3′ end formation. Nucleic Acids Res 35:3676–3692
Gigolashvili T, Yatusevich R, Berger B, Muller C, Flugge UI (2007) The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J 51:247–2461
Grubb CD, Abel S (2006) Glucosinolate metabolism and its control. Trends Plant Sci 11:89–100
Gruer MJ, Artymiuk PJ, Guest JR (1997) The aconitase family: three structural variations on a common theme. Trends Biochem Sci 22:3–6
Halkier BA, Gershenzon J (2006) Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 57:303–333
Hirai MY, Sugiyama K, Sawada Y et al (2007) Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc Natl Acad Sci USA 104:6478–6483
Knill T, Schuster J, Reichelt H, Gershenzon J, Binder S (2008) Arabidopsis branched-chain aminotransferase 3 functions in both amino acid and glucosinolate biosynthesis. Plant Physiol 146:1028–1039
Kohlhaw GB (2003) Leucine biosynthesis in fungi: entering metabolism through the back door. Microbiol Mol Biol Rev 67:1–15
Koop HU, Steinmuller K, Wagner H, Rossler C, Eibl C, Sacher L (1996) Integration of foreign sequences into the tobacco plastome via polyethylene glycol-mediated protoplast transformation. Planta 199:193–201
Kroymann J, Textor S, Tokuhisa JG, Falk KL, Bartram S, Gershenzon J, Mitchell-Olds T (2001) A gene controlling variation in Arabidopsis glucosinolate composition is part of the methionine chain elongation pathway. Plant Physiol 127:1077–1088
Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53:247–259
Roth M (1971) Fluorescence reaction for amino acids. Anal Chem 43:880–882
Sambrook J, Russel DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Sarwar G, Botting HG (1993) Evaluation of liquid chromatographic analysis of nutritionally important amino acids in food and physiological samples. J Chromatogr 615:1–22
Schuster J, Knill T, Reichelt H, Gershenzon J, Binder S (2006) BRANCHED-CHAIN AMINOTRANSFERASE 4 is part of the chain elongation pathway in the biosynthesis of methionine-derived glucosinolates in Arabidopsis. Plant Cell 18:1–16
Sessions A, Burke E, Presting G et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14:2985–2994
Singh BK (1999) Biosynthesis of valine, leucine and isoleucine. In: Singh BK (ed) Plant amino acids: biochemistry and biotechnology. Marcel Dekker, New York, pp 227–247
Sonderby IE, Hansen BG, Bjarnholt N, Ticconi C, Halkier BA, Kliebenstein DJ (2007) A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PLoS ONE 2:e1322
Strassman M, Ceci LN (1963) Enzymatic formation of alpha-isopropylmalic acid, an intermediate in leucine biosynthesis. J Biol Chem 238:2445–2452
Sureshkumar S, Todesco M, Schneeberger K, Harilal R, Balasubramanian S, Weigel D (2009) A genetic defect caused by a triplet repeat expansion in Arabidopsis thaliana. Science 323:1060–1063
Textor S, de Kraker J-W, Hause B, Gershenzon J, Tokuhisa JG (2007) MAM3 catalyzes the formation of all aliphatic glucosinolate chain lengths in Arabidopsis. Plant Physiol 144:60–71
The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
Underhill EW, Wetter LR, Chisholm MD (1973) Biosynthesis of glucosinolates. Biochem Soc Symp 38:303–326
Wittstock U, Halkier BA (2002) Glucosinolate research in the Arabidopsis era. Trends Plant Sci 7:263–270
Xing RY, Whitman WB (1991) Characterization of enzymes of the branched-chain amino acid biosynthetic pathway in Methanococcus spp. J Bacteriol 173:2086–2092
Yasutake Y, Yao M, Sakai N, Kirita T, Tanaka I (2004) Crystal structure of the Pyrococcus horikoshii isopropylmalate isomerase small subunit provides insight into the dual substrate specificity of the enzyme. J Mol Biol 344:325–333
Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3:e1994
Acknowledgments
We are grateful to Bärbel Weber, Angelika Müller and Conny Guha for excellent technical assistance. We also thank Christian Jonietz, Angela Hölzle, Daniela Köhler and Anna Rapp for their support and Matthias Schöttner for LC-TOF-MS measurements. This work is supported by DFG grant Bi 590/9-1 and the Max Plank Society.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Knill, T., Reichelt, M., Paetz, C. et al. Arabidopsis thaliana encodes a bacterial-type heterodimeric isopropylmalate isomerase involved in both Leu biosynthesis and the Met chain elongation pathway of glucosinolate formation. Plant Mol Biol 71, 227–239 (2009). https://doi.org/10.1007/s11103-009-9519-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-009-9519-5