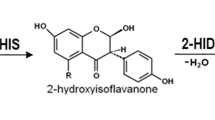
Abstract
Previous studies have identified two distinct O-methyltransferases (OMTs) implicated in isoflavonoid biosynthesis in Medicago species, a 7-OMT methylating the A-ring 7-hydroxyl of the isoflavone daidzein and a 4’-OMT methylating the B-ring 4′-hydroxyl of 2,7,4′-trihydroxyisoflavanone. Genes related to these OMTs from the model legume Medicago truncatula cluster as separate branches of the type I plant small molecule OMT family. To better understand the possible functions of these related OMTs in secondary metabolism in M. truncatula, seven of the OMTs were expressed in E. coli, purified, and their in vitro substrate preferences determined. Many of the enzymes display promiscuous activities, and some exhibit dual regio-specificity for the 4′ and 7-hydroxyl moieties of the isoflavonoid nucleus. Protein structure homology modeling was used to help rationalize these catalytic activities. Transcripts encoding the different OMT genes exhibited differential tissue-specific and infection- or elicitor-induced expression, but not always in parallel with changes in expression of confirmed genes of the isoflavonoid pathway. The results are discussed in relation to the potential in vivo functions of these OMTs based on our current understanding of the phytochemistry of M. truncatula, and the difficulties associated with gene annotation in plant secondary metabolism.
Similar content being viewed by others
Abbreviations
- EST:
-
expressed sequence tag
- IFS:
-
isoflavone synthase
- MeJA:
-
methyl jasmonate
- OMT:
-
O-methyltransferase
- TC:
-
tentative consensus
- YE:
-
yeast elicitor
References
Akashi T, Sawada Y, Aoki T, Ayabe S-I (2000) New scheme of the biosynthesis of formononetin involving 2,7,4′-trihydroxyisoflavanone but not daidzein as the methyl acceptor. Biosci Biotechnol Biochem 64:2276–2279
Akashi T, Sawada Y, Shimada H, Sakurai N, Aoki T, Ayabe S (2003) cDNA cloning and biochemical characterization of S-adenosyl-L-methionine: 2,7,4′-trihydroxyisoflavanone 4′-O-methyltransferase, a critical enzyme of the legume isoflavonoid phytoalexin pathway. Plant Cell Physiol 44:103–112
Bisby FA, Buckingham J, Harborne JB (Eds) (1994) Phytochemical dictionary of the Leguminosae, Vol I. Plants and their constituents. Chapman and Hall, New York
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Deavours BE, Dixon RA (2005) Metabolic engineering of isoflavonoid biosynthesis in alfalfa. Plant Physiol 138:2245–2259
Dixon RA, Steele CL (1999) Flavonoids and isoflavonoids- a gold mine for metabolic engineering. Trends Plant Sci 4:394–400
Edwards R, Dixon RA (1991) Isoflavone O-methyltransferase activities in elicitor-treated cell suspension cultures of Medicago sativa. Phytochemistry 30:2597–2606
Ferrer J-L, Zubieta C, Dixon RA, Noel JP (2005) Crystal structure of alfalfa caffeoyl coenzyme A 3-O-methyltransferase. Plant Physiol 137:1009–1017
Frick S, Kutchan TM (1999) Molecular cloning and functional expression of O-methyltransferases common to isoquinoline alkaloid and phenylpropanoid biosynthesis. Plant J 17:329–339
Frick S, Ounaroon A, Kutchan TM (2001) Combinatorial biochemistry in plants: the case of O-methyltransferases. Phytochemistry 56:1–4
Gang DR, Lavid N, Zubieta C, Chen F, Beuerle T, Lewinsohn E, Noel JP, Pichersky E (2002) Characterization of phenylpropene O-methyltransferases from sweet basil: facile change of substrate specificity and convergent evolution within a plant O-methyltransferase family. Plant Cell 14:505–519
Harborne JB (Eds.), (1994) The Flavonoids, advances in research since 1986. Chapman & Hall, London
Harrison MJ, Dixon RA (1993) Isoflavonoid accumulation and expression of defense gene transcripts during the establishment of vesicular arbuscular mycorrhizal associations in roots of Medicago truncatula. Mol Plant-Microbe Interact 6:643–654
He X-Z, Dixon RA (1996) Affinity chromatography, substrate/product specificity and amino acid sequence analysis of an isoflavone O-methyltransferase from alfalfa (Medicago sativa L.). Arch Biochem Biophys 336:121–129
He X-Z, Dixon RA (2000) Genetic manipulation of isoflavone 7-O-methyltransferase enhances the biosynthesis of 4′-O-methylated isoflavonoid phytoalexins and disease resistance in alfalfa. Plant Cell 12:1689–1702
He X-Z, Reddy JT, Dixon RA (1998) Stress responses in alfalfa (Medicago sativa L.) XXII. cDNA cloning and characterization of an elicitor-inducible isoflavone 7-O-methyltransferase. Plant Mol Biol 36:43–54
Hoffmann L, Maury S, Bergdoll M, Thion L, Erard M, Legrand M (2001) Identification of the enzymatic active site of tobacco caffeoyl-coenzyme A O-methyltransferase by site-directed mutagenesis. J Biol Chem 276:36831–36838
Ibrahim RK, De Luca V, Khouri H, Latchinian L, Brisson L, Charest PM (1987) Enzymology and compartmentation of polymethylated flavonol glucosides in Chrysosplenium americanum. Phytochemistry 26:1237–1245
Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta. Crystallogr A49:148–157
Joshi CP, Chiang VL (1998) Conserved sequence motifs in plant S-adenosyl-L-methionine-dependent methyltransferases. Plant Mol. Biol 37:663–674
Kim DH, Kim B-G, Lee Y, Ryu JY, Lim Y, Hur H-G, Ahn JH (2005) Regiospecific methylation of naringenin to ponciretin by soybean O-methyltransferase expressed in Escherichia coli. J Biotech 119:155--162
Kornblatt J, Muzac I, Lim Y, Ahn JH, Ibrahim RK (2004) Role of serine 286 in cosubstrate binding and catalysis of a flavonol O-methyltransferase. Biochem. Cell Biol 82:531–537
Laskowski RA, MacArthur MW, Moss DS, Throrton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures J. Appl. Cryst 26:283–291
Liu C-J, Deavours BE, Richard SB, Ferrer J-L, Dixon RA, Noel JP (2005) Dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses. Plant Cell (in review)
Liu C-J, Dixon RA (2001) Elicitor-induced association of isoflavone O-methyltransferase with endomembranes prevents formation and 7-O-methylation of daidzein during isoflavonoid phytoalexin biosynthesis. Plant Cell 13:2643–2658
Marti-Renom MA, Stuart A, Fiser A, Sanchez R, Melo F, Sali A (2000) Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct 29:291–325
Maxwell CA, Harrison MJ, Dixon RA (1993) Molecular characterization and expression of alfalfa isoliquiritigenin 2′-O-methyltransferase, an enzyme specifically involved in the biosynthesis of an inducer of Rhizobium meliloti nodulation genes. Plant J 4:971–981
Noel JP, Dixon RA, Pichersky E, Zubieta C, Ferrer J-L (2003) Structural, functional, and evolutionary basis for methylation of plant small molecules. Rec Adv Phytochemistry 37:37–58
Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. In: Carter CWJ, Sweet RM (Eds) Methods in enzymology: macromolecular crystallography, part A., Vol 276. Academic Press, New York, pp 307–326
Paiva NL, Oommen A, Harrison MJ, Dixon RA (1994) Regulation of isoflavonoid metabolism in alfalfa. Plant Cell Tissue Organ Cult 38:213–220
Phillips DA, Kapulnik Y (1995) Plant isoflavonoids, pathogens and symbionts. Trends Microbiol 3:58–64
Preisig CL, Matthews DE, VanEtten HD (1989) Purification and characterization of S-adenosyl-L-methionine:6a-hydroxymaackiain 3-O-methyltransferase from Pisum sativum. Plant Physiol 91:559–566
Schroder G, Wehinger E, Lukacin R, Wellmann F, Seefelder W, Schwab W, Schroder J (2004) Flavonoid methylation: a novel 4′-O-methyltransferase from Catharanthus roseus, and evidence that partially methylated flavanones are substrates of four different flavonoid dioxygenases. Phytochemistry 65:1085–1094
Schroder G, Wehinger E, Schroder J (2002) Predicting the substrates of cloned plant O-methyltransferases. Phytochemistry 59:1–8
Suzuki H, Reddy MSS, Naoumkina M, Aziz N, May GD, Huhman DV, Sumner LW, Blount JW, Mendes P, Dixon RA (2005) Methyl jasmonate and yeast elicitor induce differential genetic and metabolic re-programming in cell suspension cultures of the model legume Medicago truncatula. Planta 220:698–707
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
VanEtten HD, Matthews DE, Smith DA (1982) Metabolism of phytoalexins. In: Bailey JA, Mansfield JW (Eds) Phytoalexins. Blackie, Glasgow, pp 180–217
Wengenmayer H, Ebel J, Grisebach H (1974) Purification and properties of a S-adenosylmethionine: isoflavone 4′-O-methyltransferase from cell suspension cultures of Cicer arietinum L. Eur J Biochem 50:135–143
Yang H, Ahn JH, Ibrahim RK, Lee S, Lim Y (2004) The three-dimensional structure of Arabidopsis thaliana O-methyltransferase predicted by homology-based modeling. J. Mol Graph Model 23:77–87
Zubieta C, He X-Z, Dixon RA, Noel JP (2001) Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nature Struct Biol 8:271–279
Zubieta C, Kota P, Ferrer J-L, Dixon RA, Noel J (2002) Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase. Plant Cell 14:1265–1277
Acknowledgements
We thank Drs Luis Marquez and Marilyn Roossinck for help with PAUP analysis, and Drs Xiaoqiang Wang and Luzia Modolo for critical reading of the manuscript. This work was supported by grants from the Oklahoma Center for the Advancement of Science and Technology Health Sciences Program to RAD, the National Science Foundation under Grant No. 0236027 to JPN and under Grant No. DBI 0109732 to RAD, and a Noble Foundation Postdoctoral Fellowship at the Salk Institute and a Laboratory Directed Research and Development Award (LDRD) at Brookhaven National Laboratory under contract with U.S. Department of Energy to C-JL. JPN is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Deavours, B.E., Liu, CJ., Naoumkina, M.A. et al. Functional analysis of members of the isoflavone and isoflavanone O-methyltransferase enzyme families from the model legume Medicago truncatula . Plant Mol Biol 62, 715–733 (2006). https://doi.org/10.1007/s11103-006-9050-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-006-9050-x