Abstract
Chronic hepatitis B virus (HBV) infection is one of the leading causes of liver cirrhosis and hepatocellular carcinoma (HCC). Current treatment strategies of HBV infection including the use of interferon (IFN)-α and nucleotide analogues such as lamivudine and adefovir have met with only partial success. Therefore, it is necessary to develop more effective antiviral therapies that can clear HBV infection with fewer side effects. RNA interference (RNAi), by which a small interfering RNA (siRNA) induces the gene silence at a post-transcriptional level, has the potential of treating HBV infection. The successful use of chemically synthesized siRNA, endogenous expression of small hairpin RNA (shRNA) or microRNA (miRNA) to silence the target gene make this technology towards a potentially rational therapeutics for HBV infection. However, several challenges including poor siRNA stability, inefficient cellular uptake, widespread biodistribution and non-specific effects need to be overcome. In this review, we discuss several strategies for improving the anti-HBV therapeutic efficacy of siRNAs, while avoiding their off-target effects and immunostimulation. There is an in-depth discussion on the (1) mechanisms of RNAi, (2) methods for siRNA/shRNA production, (3) barriers to RNAi-based therapies, and (4) delivery strategies of siRNA for treating HBV infection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Hepatitis B virus (HBV) infection is an important global health problem with over 350 million HBV carriers worldwide (1). Approximately one million deaths occur annually due to HBV-induced liver diseases, which ranges from acute to chronic infection and from cirrhosis to hepatocellular carcinoma (HCC) (2,3). Chronically infected patients have been treated with interferon-α (IFN-α) and nucleoside analogs such as lamivudine or adefovir. IFN-α has an immunomodulatory mechanism of action and the nucleoside analogues interfere with viral DNA replication. These drugs are partially effective. Moreover, prolonged therapy with lamivudine is associated with an increased incidence of viral resistance. The low efficacy, undesirable side-effects, and occurrence of resistance to HBV mutations remain the major obstacles in their clinical application in treating HBV infection (4–7). The need for alternative therapeutic approaches has provided the impetus to develop novel therapeutic reagents for inhibiting HBV replication. Ribozymes, antisense oligodeoxynucleoitdes (ODNs) and DNA vaccines are being developed to potentially complement existing antiviral therapies (8–11).
RNA interference (RNAi) is a natural process by which small interfering RNA (siRNA) duplex directs sequence-specific post-transcriptional silencing of homologous genes by binding to its complementary mRNA and triggering its elimination (11). It is an evolutionary mechanism for protecting the genome against invasion by mobile genetic elements such as transposons and viruses. This finding generated huge interest in applying RNAi for biomedical research. Nomination of RNAi as the “Breakthrough of the year 2002” by the Journal Science prompted biologists to overhaul their vision of the cell and its evolution, and discovery of RNAi was awarded Nobel Prize to Andrew Z. Fire and Craig C. Mello in 2006.
Potent knockdown of a gene of interest with high sequence specificity makes RNAi a powerful tool for studying gene function and may be used for treating a variety of diseases. RNAi can specially inhibit the function of any chosen target genes and has shown antiviral effect against hepatitis B and C virus (HBV, HCV) and human immunodeficiency virus (HIV) (12–15). Unlike HCV and HIV, there is only modest sequence heterogeneity among HBV DNA strains. Thus, HBV makes extensive use of overlapping reading frames (ORFs) with the DNA genome, suggesting the multiple HBV RNAs will make the virus susceptible for RNA interference (16,17).
HEPATITIS B VIRUS BIOLOGY AND INFECTION
HBV belongs to a family of small enveloped DNA viruses that predominantly infects the liver. HBV particle virions are 42 nm in diameter and possess an isometric nucleocapsid core of 27 nm in diameter, surrounded by an outer coat of approximately 4 nm thick (Fig. 1). The protein of the virion coat is known as hepatitis B surface antigen (HBsAg), which consists of three subspecies, depending on the hepadnavirus. The three envelope components are called as S, M, and L (18). S protein is 226 amino acids long and known as the S domain. The two larger proteins contain S plus amino-terminal extensions of S created by initiation at upstream start codons. For HBV, these codons are located at approximately 165 (M) and 489 (L) nucleotides upstream of the initiation codon for S. The extra domain of M is known as pre-S2, while the domain unique to L is called pre-S1. S, L, and M are all found as components of the 42-nm diameter infectious viral particles (19). HBsAg is generally produced in vast excess, and is found in the blood of infected individuals in the form of filamentous and spherical particles.
Diagrammatic representation of hepatitis B virion. This virion is of spherical shape with a diameter of 42 nm, and consists of an outer envelope and an icosahedral nucleocapsid core. The outer envelope contains embedded proteins known as hepatitis B surface antigens (HBsAg). The HBV DNA genome is double-stranded and contains a long and short segment which overlap approximately 240 nt to form an open circle. The longer strand is 3020–3320 nucleotides long, and the shorter is 1700–2800 nucleotides long. The virus can be divided into four major serotypes (adr, adw, ayr, ayw) based on antigenic epitopes present on HBsAg, and into eight genotypes (A-H) according to overall nucleotide sequence variation of the genome.
HBV has a compact genome with a partially double–stranded DNA of approximately 3200 bases in length and contains four open reading frames (ORFs) that encode precore/core, polymerase, surface and HBVx (HBx) proteins. The core and polymerase genes are essential for viral DNA replication. The core gene encodes the viral capsid protein, known as hepatitis B virus core antigen (HBcAg). Additional two gene expression products are hepatitis X protein (HBx) and hepatitis e antigen (HBeAg). HBx is dispensable for virus replication in transfected cells (20,21) and acts as a stabilizer for viral RNAs (22). Apart from transactivation of many promoters, activities linked to HBx include stimulation of signal transduction (23) and binding to well-known protein targets such as p53 (24–26), proteasome subunits (27,28), and UV-damaged DNA binding protein (29). HBeAg is also dispensable for in vivo infections (30,31). HBeAg is an important marker of HBV replication and may cause depletion of Th1 helpers cells, thereby suppressing the ability to mount a strong cytotoxic T-lymphocyte (CTL) response to the infected hepatocytes (32,33).
HBV replication includes entry, uncoating, and delivery of the viral genome into the cell nucleus. The life cycle of HBV is characterized by the synthesis of a 3 kb partially double-stranded, relaxed-circular DNA (rcDNA) genome by reverse transcription of an RNA intermediate, the pregenome. The mechanism of RNA-directed DNA synthesis has been well characterized through genetic and biochemical studies (34,35).
All hepatitis viral transcripts utilize a common polyadenylation signal located within the core protein-coding region (Fig. 2). The 3.5 kb pregenomic RNA and three subgenomic mRNAs regulate the translation of HBV proteins and reverse transcription of HBV DNA (18). The pregenomic mRNA not only serves for translation of the core protein, HBeAg and polymerase-reverse transcriptase, but also represents the template for reverse transcription. The DNA polymerase and X gene are essential for reverse-transcription and transcription regulation. Both the life cycle and genome structure of HBV are susceptible to specific siRNAs.
A schematic diagram depicting the location of siRNAs in association with viral open reading frames and viral mRNAs within the HBV genome. Downward red arrows indicate the location of RNAi target sites within the four HBV transcripts, the siRNA4 targets to the common HBV polyadenylation signal region. The 3.5 kb transcript is the pregnomic RNA that serves as the template for HBV DNA replication and encodes the viral core and polymerase protein. 2.4 kb and 2.1 kb mRNA encode the viral envelope proteins. 0.7 kb mRNA encodes the viral X protein.
MECHANISMS OF RNA INTERFERENCE
RNAi is a regulatory mechanism of most eukaryotic cells that uses dsRNA molecules to direct homology-dependent control of gene activity (36). RNAi-based gene silencing occurs post-transcriptionally in the cytoplasm (37) and it is an ATP-dependent and translation-independent event. The RNA-induced silencing complex (RISC) model is showed in Fig. 3. siRNAs are 9–24 bp dsRNA molecules and have a characteristic 2 nucleotide 3′-overhang that allows them to be recognized by the enzymatic machinery of RNAi, leading to homology dependent degradation of the target mRNA. In mammalian cells siRNAs are produced from cleavage of long dsRNA precursors by the RNase III endonuclease Dicer (38). Dicer works with a small dsRNA-binding protein, R2D2, to hand off the siRNAs to the RISC, which contains the “splicing” protein Argonaute 2 (Ago2) that cleaves the target mRNA molecules between bases 10 and 11 relative to the 5′-end of the antisense siRNA strand. The core components of RISC are the Ago family members, and in humans only Ago2 possesses an active catalytic domain for cleavage activity (39). While siRNAs loaded into RISC are double-stranded, Ago2 cleaves and releases the “passenger” strand leading to an activated form of RISC with a single stranded “guide” RNA molecule (40). mRNA molecules with perfectly or near-perfectly complementary to the guide RNA are recognized and then cleaved by Ago2.
Mechanism of RNA interference (RNAi). Only core components of the RISC Loading Complex (RLC) and RISC are depicted. RLC contains a Dcr-2/R2D2 heterodimer which binds the siRNA containing dinucleotide 3’-overhangs. Core RISC component Ago2 displaces Dcr-2/R2D2. This schematic depicts transfer of duplex siRNA to Ago2 with either concurrent or immediate Ago2-mediated cleavage of the passenger strand. ATP hydrolysis is required for RISC maturation and has been postulated to accelerate release of the cleaved passenger strands as it does for cleaved mRNA. Mature RISC guides strand of the siRNA, and cleave mRNA targets. The pink arrows represent the pathway for a shRNA (A). The green arrows represent the pathway for a long dsRNA (B). The yellow arrows represent the pathway for a siRNA (C).
A short hairpin RNA (shRNA) is a DNA-based siRNA production strategy in which siRNAs are produced by intracellular processing of shRNA transcripts (Figs. 3 and 4). shRNA uses a plasmid or viral vector and utilizes the H1 or U6 promoter to facilitate shRNA expression. The shRNA hairpin structure is cleaved by the cellular machinery into siRNA, which is then bound to the RISC. This complex binds and then cleaves mRNAs which match the siRNA that is bound to it. The shRNA is processed in the cytoplasm by the RNAse III Dicer and one of the two strands is loaded into RISC in the cytoplasm, the loop is removed.
OFF-TARGET EFFECT
siRNA duplexes can potentially target multiple genes in addition to the intended target. This phenomenon is known as “Off-target activity”, which can principally arise from two mechanisms: depletion on the mRNA level or the suppression of translation at protein level. Alternatively, another possible mechanism for off-target activity arises from the fact that the physical structure of siRNAs appears to be identical to the related class of miRNAs. miRNAs seem to have mismatches between RNA oligo and RNA target inherent in their structure. A siRNA can potentially interact with the 3′-UTRs of unintended genes that yield mRNA translation inhibition rather than mRNA degradation. So, off-target effects of siRNAs is reminiscent of transcript regulation by miRNA (41). Therefore, improved understanding of the miRNA mechanism of action could also allow us to improve the design of siRNAs and to improve the design of experiments where siRNAs are used. Recently, 2′-O-methyl (2′-OMe) modifications to specific positions within a siRNA seed region reduced both the number of off-target transcripts and the magnitude of their regulation as well as off-target phenotypes in functional study, without significantly affecting silencing of the intended targets (42).
siRNA delivery into mammalian cells often induces innate interferon (IFN) responses and sequence-independent suppression of both endogenous and exogenous gene expression into “off-target” effect. The use of siRNA containing immunostimulatory RNA motif or siRNA with length of 30 bp or more can be complicated due to the induction of the nonspecific IFN response in mammalian cells through the RNA-activated protein kinase (PKR) and interaction with endosomal Toll-like receptors (TLRs), in particular TLR7/8. This effect may be of greater significance when we use perfectly matched exogenous RNA for gene silencing (43–45) or when we select cationic liposomes or polymers for siRNA delivery (46). TLR7 binding is sequence specific, favoring GC-rich sequences, and can be avoided by choosing sequences that are not recognized by this receptor. Moreover, this effect leads to the production of proflammatory cytokines and type I infections and induce nuclear KB activation. 2′-Uridine modifications within the siRNA duplex not only evades immune activation, but also suppresses TLR signaling triggered by their unmodified counterparts, 2′-OMe modified siRNAs are recognized with high affinity by TLR7/8, but do not induce downstream signaling (47). These off-target activities potentially cause the inhibition of translation and mRNA degradation of unintended genes, which would be undesirable in a therapeutic process.
DELIVERY STRATEGIES
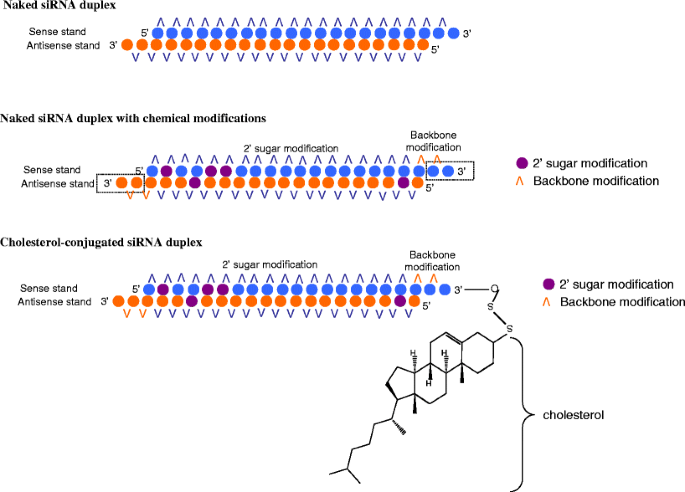
To improve the effect of RNAi-based therapy, the enzymatic stability and cellular uptake of siRNA should be significantly enhanced, while their immunoactivation should be decreased. Chemical modification and bioconjugation with lipids are known to improve the stability and cellular uptake of siRNA. The overall scheme for improving siRNA stability, cellular uptake and gene silencing efficiency is illustrated in Fig. 5.
Scheme for improving siRNA stability, cellular uptake and gene silencing. Important features of siRNA structures include two base pair overhangs, seed region and mRNA cleavage site. A siRNA duplex with phosphorothioate (P=S) and 2’-base sugar modification (purple circle) confer resistance against exonuclease and endonuclease in comparison with a naked siRNA duplex. Bioconjugation with cholesterol or carrier molecules carrying targeting ligands increases cellular uptake and site-specificity.
Chemical Modification
The hydrophilic character and anionic backbone of siRNAs reduces their uptake by the cells. While siRNA duplexes are relatively more stable in serum than single stranded siRNA, they easily undergo degradation in vivo (48–51). This represents a significant barrier to the therapeutic development of siRNA. Chemical modifications to sugars, backbones, or bases of siRNAs, have been shown to enhance their stability, prevent them from triggering an immune response, control their pharmacokinetic profiles and reduce nonspecific effects without affecting their biological activity. Most commonly used chemical modifications of siRNA include phosphorothiolation (P=S) of the non-bridging oxygen at the 3′-end, 2′-sugar modification (such as 2′-OMe or 2′-F) and locked nucleic acid (LNA). Chemical modifications of siRNA confer enhanced gene silencing at lower doses and reduced dosing frequency (52). siRNA molecules consisting of 2′-OMe and 2′-F have shown to have enhanced stability in the plasma and increased in vivo potency (48,49). The modifications also reduce the immunostimulatory effect of siRNAs (53). Judge et al. reported that immune stimulation by synthetic siRNA can be completely abrogated by selective incorporation of 2′-OMe uridine or guanosine nucleosides into one strand of the siRNA duplex. These non-inflammatory siRNAs, which contain less than 20% modified nucleotides, can be readily generated without disrupting their gene-silencing activity (54,55). Morrissey et al. (56) selected two siRNAs, HBV263 and HBV1583 and chemically stabilized for nuclease resistance using 2′F, 2′-OMe or 2′H residues to substitute 2′-OH residues. They then analyzed in vitro potency (Fig. 6). It was found that the placement of one, two or three ribonucleotides at the 5′ end of the modified siRNA improved the median inhibitory concentration of the stabilized siRNAs by approximately fivefold. Jackson et al. (2007) demonstrated that 2′-OMe modification of position 1 and 2 within the siRNA seed region reduce both the number of off-target transcripts and the magnitude of their regulation, without significantly affecting silencing of the intended target (42). 2′-OMe modification at the 5′-end significantly prolongs the half-life of a large fraction of the siRNA guide strand when exposed to human blood serum (49). Furthermore, siRNA with 2′-OMe modification completely block ribonucletic cleavage 3′ to position 1. It also significantly reduces but does not completely block cleavage 3′ to position 2.
HBV-specific siRNAs and their in vitro potency. (A) Chemical modifications of HBV-specific siRNAs. (B) Activity of chemical modified siRNAs incorporated with stable nucleic acid lipid particles (SNALP) against HBV in cell culture system. Secreted HBsAg levels were assayed by ELISA from HepG2 cells transfected with HBV expression vector and subsequently treated with HBV263M-SNALP, HBV1583M-SNALP, HBV263invM-SNALP, or an HCV irrelevant control (HCVirrM) at final concentrations of 0.5 to 25 nM. HBsAg levels were assayed 3 days after transfection, and expressed as OD 450 nm. Reproduced with permission from Morrissey et al. (2004).
Bioconjugation
An siRNA can be linked to a carrier molecules or covalently conjugated to a targeting ligand via a cleavable spacer to increase its cellular uptake or confer cell-specific targeting. Since the antisense strand is active, the carrier molecule is usually attached to the sense strand of an siRNA duplex without compromising the level of gene silencing. Conjugates can be placed on either the 5′- or 3′-end of the sense strand. To date, siRNA has seen conjugated to lipids, polymers, peptides and aptamers (57–60).
Apolipoprotein B (ApoB) is expressed predominantly in the liver cells, and is an essential protein for assembly and secretion of very-low-density lipoprotein (VLDL) and low-density lipoprotein (LDL), which are required for the transport and metabolism of cholesterol. Soutschek et al. conjugated cholesterol to the 3′-end of the sense strand of siRNA targeting ApoB and demonstrated significant increase in the levels of this conjugate in the liver. Most importantly, siRNAs efficiently reduced the levels of ApoB mRNA in the liver by ∼55% and ∼70% in the Jejunun, the two main sites of AboB expression. Concomitantly, ApoB in plasma protein dropped by ∼70% and serum cholesterol fell by 35–40% (53). Lorenz et al. also have conjugated siRNAs to lipids like lithocholic and lauric acids at the 5′-end of the sense strand and showed increased cellular uptake of siRNAs in human liver cells without the use of any transfection reagent (61).
Conjugation of siRNA to apatamer which can bind to prostate-specific membrane antigen (PSMA) has shown some promise for targeted delivery of siRNA to prostate cancer. McNamara et al. developed aptamer-siRNA chimeric RNAs capable of cell type –specific binding and delivery of functional siRNA into cells. The aptamer portion of the chimeras mediates binding to PSMA, whereas the siRNA portion targets the expression of survival genes when applied to cell expressing PSI (57,62).
Complex Formation
Cationic liposomes have been widely used for transfection of siRNA and shRNA. Gene silencing efficiency greatly depends on the cationic lipid structure and liposome composition (63). The cationic lipid which contains protonable polyamines linked to dialkyl or cholesterol anchors in the liposomal formulation form complexes with DNA or RNA, leading to their enhanced cellular association. Morrissey et al. (56,64) incorporated chemically modified siRNA against HBV into a specialized liposome to form a stable nucleic acid lipid particle (SNALP) and administered intravenously into mice carrying replicating HBV. There was significant improvement in efficacy compared to unformulated siRNA with a longer half-life in plasma and liver. Mahato et al. (65) reported that cationic lipids with multivalent head-groups are more effective in gene transfer compared to their monovalent counterparts.
Hepatocytes can be targeted using galactosylated liposomes (66–68) or asialofetuin grafted vesicles (69). Liposomes that are modified with human serum albumin predominantly accumulate in liver endothelial cells (70) and mannosylated liposomes are recognized by Kupffer cells (71,72). To deliver antisense ODN into hepatocytes for treating HBV infection, Zhang et al. (73) developed the liver-targeting cationic liposomes as a gene carrier, which was co-modified with the ligand of the asialoglycoprotein receptor (ASGPR), beta-sitosterol-beta-d-glucoside (sito-G) and the nonionic surfactant, Brij 35. The cationic liposomes/ODN complexes exhibited high transfection efficiency and strong antigen inhibitory effect in primary rat hepatocytes and HepG2.2.15 cells, respectively. The cellular uptake of these liposomal formulations is mediated by endocytosis and membrane fusion. The ligand sito-G enhanced ASGPR-mediated endocytosis, while the nonionic surfactant Brij 35 facilitated membrane fusion. This co-modification resulted in efficient transfection, with little cytotoxicity. These results suggest that the co-modified liver-targeting cationic liposomes would be effective to ODN delivery to hepatocytes infected with HBV.
Like cationic lipid, cationic polymer can also form complexes with siRNA because of electrostatic interaction between positively charged amine groups of the polycations and negatively charged phosphate groups of the siRNA. The interaction between cationic polymer/siRNA complexes and negatively charged cell membranes can enhance its cellular uptake and thus increase transfection efficiency (74). Various cationic polymers have been studied as potential vectors for gene delivery which include proteins such as histones and cationized human serum albumin, polypeptides such as poly-l-lysine (PLL) and poly-l-ornithine, and polyamines such as polyethyleneimine (PEI) and starburst polyamidoamine (pAMAM) dendrimers. pAMAM dendrimers have been largely applied for delivery of plasmids and ODNs (75,76), but not yet explored for siRNA delivery. Generally, it is expected that the advantages observed for ODNs may be adapted for siRNA. Among the cationic polymers employed for gene delivery, PEI has been the most widely used polymer for siRNA delivery. PEIs with various molecular weights, degrees of branching, and other modifications have been used for transfecting siRNAs in different cell lines and live animals (77,78). The high transfection efficiency of PEI can be attributed to its buffering effect, also known as “proton sponge effect” due to its secondary and tertiary amines. The cytotoxicity and transfection efficiency of PEI are directly proportional to its molecular weight. PEI is often grafted with polyethylene glycol (PEG) or cholesterol to reduce its cytotoxicity (79,80).
Antibody-mediated delivery is an effective method of targeting siRNA to particular cells. This involves fusing the nucleic acid binding domain to Fab fragment or scFv that recognizes a membrane receptor, resulting in a fusion protein that possesses cell recognition and nucleic acid-binding abilities. This fusion protein can then bind nucleic acids and deliver them into target cells through receptor-mediated endocytosis. Wen et al. (81) recently tested five siRNA duplexes to select a specific siRNA that effectively inhibited HBV gene expression and replication. These investigators constructed two fusion proteins, s-tP and sCkappa-tP to contain a single chain of the human variable fragment, scFv, against HBsAg, a truncated protamine (tP). To provide targeted delivery of siRNA, siRNA expressing cassettes (SEC), and siRNA-producing plasmids, a constant region of the kappa chain (Ckappa), s-tP and sCkappa-tP were developed. Fluorescein labeled-siRNA, fluorescein labeled-SEC, and plasmid DNA were specifically delivered into HBsAg-positive cells using the sCkappa-tP fusion protein, and effectively inhibited HBV gene expression and replication. HBV gene expression was also inhibited by siRNA or siRNA-producing plasmids in HBV transgenic mice.
Hydrodynamic Injection
The hydrodynamic injection technique is used to efficiently deliver HBV expression plasmids and siRNAs targeting various HBV gene regions, with enhanced HBV gene silencing (64,82,83). Morrissey et al. (64) assessed the activity of stabilized siRNAs in vivo after co-delivering an HBV vector and siRNA via high-volume tail vein injection. More than a 3 log10 decrease in levels of serum HBV DNA and HBsAg, as well as liver HBV RNA, were observed in the siRNA-treated groups compared to the control siRNA-treated and saline groups. Furthermore, the observed decrease in serum HBV DNA was 1.5 log10 more with stabilized siRNA compared with unmodified siRNA, indicating the value of chemical modification in therapeutic applications of siRNA.
METHODS OF siRNA PRODUCTION
siRNAs can be produced either by chemically synthesis or by in vitro transcription or in vivo vector expression of shRNA.
Chemical Synthesis of siRNA
The selection of siRNA for a target gene is a crucial step for its application, yet empirical process is used for selection of siRNA because the rules that govern efficient siRNA silencing are still unknown. Nevertheless, Ambion (Houston, TX) recommended four guidelines for designing siRNAs: (1) beginning with the AUG of the target gene transcript, search downstream for AA dinucleotide sequences, each AA and the 3′ adjacent 19 nt are potential siRNAs; (2) blast the potential sequences against the species-specific genome database to eliminate cross-silencing phenomenon with non-target genes; (3) select three to four target sequences along the gene for production of siRNAs; and (4) for all siRNA experiments, negative control siRNAs with the same nucleotide, but a scrambled sequence (84). Ui-Tei et al. (85) proposed another four practical guidelines for designing siRNA sequences in mammalian cells: (1) A/U at the 5′-end of the antisense strand; (2) G/C at the 5′-end of the sense strand; (3) at least five A/U residues in the 5′ terminal of the third antisense strand; and (4) the absence of any GC stretch of >9 nt in length. These guidelines closely related to the molecular mechanism of RISC assembly.
Web-based software and bioinformatics is the current method for siRNA design. Naito et al. (86) presented a web-based software, siDirect (http://design.RNAi.jp) (86), to compute highly effective siRNA sequence with maximum specificity. The siDirect algorithm incorporated the four guidelines to favor efficient mammalian RNAi with a high success rate. In addition, it investigates all the potential cross-hybridization to avoid off-target gene silencing effects. Bioinformatics approaches have also been used to help select optimal siRNAs. Yuan et al. (87) built a web-based tool (http://jura.wi.mit.edu/bioc/siRNA) that implements several algorithms to identify siRNAs with high probability of silencing the target gene. One advantage of the server is that they keep incorporating rules from new results, which provides the biologist access to the newest design features. To design esiRNAs, Henschel et al. (88) developed a web-based tool, DEQOR(http://cluster-1.mpi-cbg.de/Deqor/deqor.html). The program mimics esiRNAs by fragmenting the input sequence into small pieces (16–25 nt), whereby the sequence window is shifted along the input query by 1 nt at each iteration step of the algorithm. Each simulated siRNA will be: (1) analyzed using a scoring system based on the state-of-the-art parameters for its ability to induce specific gene silencing; and (2) analyzed for its ability to cross-silence genes different from the target by performing BLAST searches against the transcriptome or genome of the studied organism. Although algorithms can increase the chances of identifying active siRNAs, they can sometimes miss the most potent siRNAs, which can only be identified by experiments testing.
Chemical synthesis is a direct means of generating siRNAs. Advantages of chemically synthesized siRNA include the precise control of the amount and purity of siRNA, ease in characterization and modification of siRNA (89), high transfection efficiency into the cells of interest, and immediate gene silencing (90,91). The steps of chemical synthesis of siRNA includes the production of sense and antisene strands, annealing of the strands, adding stable chemical entities and 2 nt overhangs at 3′ end. Dykxhoorn et al. (90) reported that siRNA duplex requires a 3′-hydroxyl group and a 5′-phosphate group for functional activity.
Although siRNA provides a powerful tool for inhibiting target genes, there are several concerns and limitations in the practical application of this technology. Milhavet et al. (84) summarized several parameters that could affect transfection and gene silencing efficacy: (1) cell culture conditions, cell density and medium composition; (2) amount and type of transfection agent; (3) quality and amount of siRNA; and (4) exposure time of the cells to siRNA. Furthermore, the use of chemically synthesized siRNA is limited by the fact that different sequences have significantly different inhibitory abilities. Holen et al. (92) observed that several siRNAs synthesized against different sites on the same target mRNA demonstrated striking different gene silencing efficiency, and only a few of the siRNAs resulted in a significant gene silencing, suggesting that accessible siRNA target sites may be rare in some human mRNAs. Hohjoh et al. (93) reported that different siRNAs induce different levels of gene silencing.
Several sites of the HBV genome have been targeted to assess the efficiency of chemically synthesized siRNAs against HBV (Table I). These include pre C/core, HBsAg and HBx. The most commonly used strategy is to use chemically synthesized double-strand siRNAs of 19–24 nt. These siRNA duplexes are usually transfected into HBV producing HepG2.2.15 or HuH7 cells. Sequence-specific reduction in viral RNA and DNA production are often seen. This accompanies by a drop in the secretion of HBsAg and/or HBeAg into the culture medium (82,94,95). Hamasaki et al. (95) demonstrated that siRNAs specific for the 3.5 kb pregenomic RNA, which did not bind to other viral transcripts reduced the level of 3.5 kb pregenomic RNA and resulted in reduced levels of secreted HBeAg and replicative intermediates converted from the 3.5 kb pregenomic RNA. Unsurprisingly, the levels of 2.4/2.1 kb mRNA and secreted HBsAg was not reduced in cells transfected with siHBV, since the 2.4/2.1 kb mRNA did not include the homologous sequence to HBV.
Because all HBV viral RNAs have a common polyadenylation signal at their 3′-ends located within the core protein-coding region, effective siRNA targeted region that can decrease the levels of 3.5 kb, 2.4 kb, and 2.1 kb HBV mRNA. Konishi et al. (94) selected the polyadenylation signal as one of the siRNA targets based on their previous successful results using oligoDNA (96). The site offers the possibility of disrupting several sensitive sites due to overlapping reading frames. The use of siRNA against HBV is attractive because of its small size mRNA and overlapping reading frames. It is possible to design siRNAs such that a single sequence could disrupt several critical mRNAs. The observed decreases in levels of 3.5-kb, 2.4-kb, and 2.1-kb HBV mRNA are consistent with this concept (Fig. 2).
RNAi target sequences should be chosen on the basis of their conservation among the major HBV genotype. The conserved gene regions of HBV genome are chosen as the target sites of siRNA sequences. Compared to RNA viruses, which have a high rate of mutation during their RNA genome replication, the genomes of DNA viruses are relatively stable, with a low mutation rate. Thus, resistance to RNAi-mediated viral gene silencing though mutation of the target sequence has not been observed in DNA virus infection. However, siRNA may exert selection pressure on preexisting resistant mutants, as exemplified by a recent report on HBV (97). Although HBV is a DNA virus, its DNA replicates through a genomic RNA intermediate and utilizes a virally encoded reverse transcriptase. Consequently, a significant amount of diversity, similar to that seen in RNA viruses, occurs in the sequences of HBV isolates, which can be classified into at least 8 genotypes (A–H). A mutant C genotype that contains a mutant in the 3′ portion of the targeted region was isolated and was more resistant to HBV S1 shRNA-mediated RNAi than the wild-type. This mutant can be preferentially selected in the presence of HBV S1 shRNA in Huh-7 cells co-transfected with wild type and mutant HBV-C at a ratio of 9:1 at day 5 post-transfection (97). Many studies have found that the distinct RNAi targeting sequences on HBsAg gene exhibited different efficacies on inhibition of the viral DNA replication and gene expression (12,82,98,99). The highest inhibitory effects induced by different siRNAs targeted on HBcAg and HBx gene have also been reported (12,98). The particular advantage of using RNAi-based therapeutic strategy over conventional drugs is that siRNA mediated inhibition of gene expression does not require any viral DNA replication.
One of the major challenges to the clinical applications of RNAi technologies is how to handle escape from inhibition of viruses that are prone to a high mutation rate and different RNAi exhibiting different degrees of gene silencing. The latter may be handled by screening effective siRNA sequences. Selection of RNAi escape mutants has been reported in vitro for poliovirus, HIV-1 and HCV. However, resistant viral sequences were susceptible to silencing by corresponding site mutant siRNAs or alternative site siRNAs. Several sites of the HBV genome have been targeted to assess the efficiency of siRNAs against HBV which include C, S, and X gene regions of HBV (100,101).
Transfection of a single siRNA often fails to provide adequate gene silencing. Another potential problem of chemically synthesized siRNA is variable transfection efficiency. To circumvent these limitations, expression vectors containing shRNA expression cassettes are developed which undergo processing by Dicer in mammalian cells. ShRNAs are generally designed to match for the target mRNA according to corresponding siRNA sequences to induce gene silencing. These shRNAs can be produced from plasmid or viral expression vectors.
Although the siRNAs sequences used by different researchers are different, a series of experiments showed significant reduction in HBV transcripts and proteins in cell culture, as well as in the viral replicative forms, induced by siRNA-producing vectors (55,98,102–105).
Endogenous Expression of Anti-HBV shRNA
Plasmid and viral vectors have been developed to express shRNA that are converted into siRNA in cells. Briefly, in the plasmid-based expression vector, the shRNA cassette including sense and antisense of siRNA sequence is transcripted as a single RNA with a short loop of 4–10 base sequence in the middle (Fig. 3). The transcript forms a hairpin structure that can be digested into functional siRNA by Dicer upon exporting into cytoplasm. Two different promoters, namely polymerase III (pol III) U6 and H1 promoters are used predominantly (106–108). It was reported that the vector containing Pol III promoter for expressing shRNA does not induce interferon responses like pol II.
One emerging concern is that RNAi mono-therapies might ultimately fail to control viruses that can escape silencing by mutation and/or RNAi suppression. Thus, sophisticated strategies are being developed that aim at averting viral resistance by combining RNAi effectors with each other or with further gene expression inhibitors. Several reports have already validated this new concept of combinatorial RNAi (coRNAi) and illustrate its versatility by co-expression of shRNA against single or multiple, viral or cellular targets. Moreover, vectors have been engineered to blend RNAi-mediated gene inhibition with conventional gene replacement strategies.
Plasmid Vector
Many studies have shown that siRNAs based on chemical synthesis could knock down the expression of HBV gene (82,94,109). However, most of the studies showed transient effects using the chemically synthesized siRNA for an infected cell line or co-transfecting a vector bearing HBV genome with siRNA to a healthy cell line. The vector-based shRNA, targeted against specific sequences in the HBV genome has been shown to inhibit the expression and replication of the HBV in vitro and in vivo.
Table II summarizes recent studies using shRNAs to target the transcripts from all four major HBV ORFs (12,38,97–99,101,102,104,105,110,111). While these studies demonstrated the feasibility of achieving efficient viral gene silencing, there was no clear pattern due to the differences in the experimental settings. Most noteworthy finding were by McCaffrey et al. (12) who demonstrated shRNAs can block replication of HBV in cell culture and in immunocompetent and immunodeficient mice transfected with plasmid encoding shRNA against HBV. These authors injected a large volume of plasmids encoding the HBV genome together with HBV-specific shRNAs into mouse liver. Among the organs transfected with the exogenous HBV DNA (liver, spleen, kidney and pancreas), the liver showed the highest levels of gene expression with approximately 40% of hepatocytes expressing the transgenes. The authors show that the HBV-specific shRNAs significantly reduced viral mRNAs and protein expression, thus inhibiting virus replication in hepatocytes of HB-infected mice.
Although shRNA expressed from the plasmid driven by polymerase III using the constitutive promoter, such as U6 and H1, were able to knock down HBV expression, most of the observations were transient. In an attempt to improve the silencing effect, Ren et al. (104) used a modified cytomegalomavirus (CMV) promoter to drive expression with Polymerase II (Pol II), which is considered to be a stronger promoter than the other common RNA Pol II promoters. These authors constructed seven vectors expressing specific shRNA driven by RNA polymerase II promoter and transfected into Hep2.2.15 cells. In the cells that can stably express functional siRNA, HBsAg and HBeAg secretion into culture media was inhibited by 86% and 91%, respectively. Immunofluorescence and Western blot analysis indicated similar results. HBV DNA was markedly restrained by 3.28-fold. RT-PCR analysis showed that viral mRNAs were effectively degraded, thus eliminating the messengers for protein expression as well as templates for reverse transcription.
Due to their short half-life and low in vivo transfection efficiency, plasmid-based shRNAs are unlikely to be effective in treating chronic HBV infection, because virtually all hepatocytes in the patients are infected. In this regard, a viral vector, which can achieve efficient and uniform transduction of all liver cells, should provide an effective gene therapy for chronic HBV infection.
Viral Vector
While most rapidly dividing cell lines are easily transfected using shRNA expression plasmids, these vectors are not easily transfected into primary cells, stem cells, and nondividing cells. In the absence of cell division, the siRNA expression plasmids cannot be introduced into the nucleus, where the DNA is transcribed. To overcome this limitation, different viral vectors encoding shRNA including lentiviral, adenoviral (Adv), and adeno-associated viral (AAV) vectors are being developed (112–115).
Uprichard et al. (99) and Carmona et al. (117) have recently demonstrated that shRNAs delivered by recombinant Adv vector could suppress HBV replication and gene expression in transgenic mice, an animal model more clinically relevant to chronic HBV in terms of containing heavily ongoing HBV replication in all hepatocytes.
AAV vectors are nonpathogenic and less immunogenic compared with other viral vectors. The ability of AAV vectors to infect both dividing and non-dividing cells, and to direct long-term gene expression in these tissues made it an ideal vehicle for gene therapy. Chen et al. (118) used a double-stranded adenoassociated virus 8-pseudotyped vector (dsAAV2/8) to deliver shRNA. HBV transgenic mice were used as an animal model to evaluate the inhibition effects of the RNAi-based gene therapy. A single administration of dsAAV2/8 vector, carrying HBV-specific shRNA, effectively suppressed the steady level of HBV protein, mRNA and replicate of DNA in the liver of HBV transgenic mice, leading to 2–3 log10 decrease in HBV concentration in the circulation. The therapeutic effect of shRNA was sequence-dependent and did not involve activation of interferon. These results underscore the potential of developing RNAi-based therapy by dsAAV2/8 vector to treat HBV chronic infection.
Sun et al. (119) selected two effective siRNAs targeting S and X gene region of HBV genome and cloned them into a human foamy virus (HFV)-based vector to generate a dual siRNA expression vector HFVU6-siSX. The results showed that the siRNA vector effectively inhibited multiple HBV gene expression and viral DNA replication.
Combination Strategy of RNAi
The ongoing solution to thwart viral mutation evolution and circumvent the resistance of anti-viral therapy is to combine RNAi triggers or with other ways of inhibiting gene expression. These novel approaches hold significant promise over conventional mono-therapy.
Cocktail of siRNAs
Variability of gene silencing efficiency of different siRNA sequences can be attributed to different affinity between cellular siRNA binding protein and the secondary and/or tertiary structures of target mRNA participating in the induction of RNAi activities (120). Using the cocktail of siRNAs could be solved the problem because the combination of siRNAs will cleave multiple sites in the mRNA target and make it difficult for the target to be repaired. For example, Chen et al. (121) demonstrated combination of siRNAs targeting different HBV sites was highly effective in the inhibition of HBV replication and antigen expression compared with the use of any individual siRNA (121). Therefore, it is important to use a cocktail treatment with the capability of inhibiting multiple viral antigen expression and DNA replication (94).
Production of Multiple siRNAs
The use of a vector which can generate multiple siRNAs or shRNAs would have the advantage of limiting escape and targeting a range of sequences found in different viral genotypes or quasispecies. It is important to use a combination of various highly effective shRNAs against S, C, P and X genes to achieve complete inhibition of HBV replication and gene expression. To toward this goal, some investigations to determine the best combination of the effective shRNAs are warranted.
Weinberg et al. (122) assessed the efficacy of long hairpin RNAs (lhRNAs) that target the conserved HBx open reading frame of HBV. As substrates for Dicer, lhRNAs have the potential to generate multiple siRNAs to enable simultaneous targeting of different sites (Fig. 7). These authors constructed two U6 Pol III vectors that encode anti-HBV lhRNAs with a 62 base pair stem sequence containing multiple G:U pairings. Assessment in transfected cultured cells and also in vivo using the hydrodynamic injection model in mice showed that one of the lhRNA vectors (lhRNA 1) decrease viral replication by 70–90% with no evidence of interferon response induction. The highest silencing efficacy was observed for targets that are complementary to sequences located at the base of the hairpin stem and this correlated with a higher concentration of siRNAs derived from this region of the lhRNA.
Long hairpin RNA (lhRNA) sequences and HBV target sites. (A) Schematic illustration of lhRNA comprising 62 bp in the stem. G:U pairings are shown as well as a sequence of 2 U residues that are derived from the transcription termination signal. The antisense strand is perfectly complementary to its HBV target. (B) Organization of the HBV genome showing open reading frames (ORFs) and sites within the target vector, which are complementary to antisense components of lhRNA. Four arrows indicate the HBV transcripts, which have common 3′-ends, and include the lhRNA targets. (C) Hepatocyte concentrations of HBV mRNA from the core and surface regions expressed as a ratio to amount of GAPDH mRNA in vivo. Total RNA was isolated from liver cells at day 5 after hydrodynamic injection and subjected to quantitative real-time PCR. (D) HBsAg secretion from Huh7 cells cotransfected with indicated hairpin RNA-encoding plasmids together with HBV target plasmid. (E) Serum HBsAg concentrations were determined at day 5 after hydrodynamic injection of mice with pCH-9/3091 HBV target and indicated hairpin encoding sequences. Reproduced with permission from Weinberg et al. (2007).
Combination of siRNA and Other Inhibitory Factors
To combine shRNA with other inhibitors of gene expression is another promising way to overcome viral gene mutation and improve the silencing effect. Jarczak et al. (123) investigated a dual strategy of ribozyme and siRNA simultaneously targeting distinct highly conserved HCV 5′- and 3′-UTR RNA sequences. Huh-mono cells were co-transfected with U6 promoter-driven siRNAs and tRNAVal-driven hairpin ribozymes with attached CTE (tRz-CTE). tRz induced 30% decrease in NS5B protein levels. siRNAs reduced HCV NS5B expression by approximately 50%. Surprisingly, when U6 promoter-driven siRNAs were combined with tRNAVal-driven hairpin ribozyme tRz-CTE, a consistent inhibitory effect of the ribozyme in the range of 25% was added to the inhibitory effect of each siRNA. This suggests that a dual strategy of ribozymes and siRNAs might become a powerful molecular tool to specifically silence HCV RNA replication.
HBV replicates through a pregenomic RNA (pgRNA) intermediate, which provides a therapeutic opportunity for a novel antiviral gene therapy based on combination of ribozyme RNA cleavage and siRNA.
Combination of siRNA and Nucleoside Analogues
Nucleoside analogues like lamivudine therapy have been shown to a rapid decrease in HBV DNA and improvements in transaminase levels and liver histology and enhanced rate of loss of HBeAg. In patients who do not lose HBeAg, stopping therapy after 3 to 12 months is usually followed up by a return of HBV DNA to pretreatment values and relapse of disease. A major limitation of chronic lamivudine therapy, however, is the development of viral resistance, marked virologically by rises in HBV-DNA levels despite continuation of this therapy. Resistance to lamivudine typically develops after 6 months of treatment and is associated with mutations in the highly conserved catalytic region of the HBV polymerase gene (124). Combination of siRNA and lamivudine may exhibit greater inhibitory effect on HBV replication. Li et al. (116) systemically measured the anti-HBV effect of siRNA combined with lamivudine on HepG2.2.15 cells at HBeAg, HBsAg, DNA and mRNA levels. The combination of siRNA and lamivudine exerted a pronounced inhibition of HBV replication.
CONCLUDING REMARKS
HBV viral infection remains a challenge for modern medicine. RNAi can inhibit HBV gene expression and replication, and it might have the potential to revolutionize the treatment of HBV. The level of HBV gene silencing greatly depends on siRNA sequences and dose. Virus mRNA may form complicated space structure which obstruct siRNA to come into play a role. A single nuclear mutant on the target gene may obstruct gene silencing. HBV DNA replicates through pre-mRNA intermediate and reverse transcriptase, HBV genome is prone to mute, and mutation strains escape silencing by RNAi. Thus, strategies should be developed that can effectively avert viral mutation by combining siRNAs which can simultaneously target multiple sites of HBV gene.
Viral gene-specific siRNAs are promising antiviral inhibitors and have been examined in a broad range of medically important viruses. However, many RNA viruses escape RNAi-mediated suppression by counteracting the RNAi machinery through mutation of the targeted region, by encoding viral suppressors, or both. DNA viruses also counteract the RNAi machinery, preferentially using viral suppressors. Cellular factors may also contribute to RNAi resistance. Systemic delivery of synthetic siRNA and shRNA expression vectors to specific targets is still a problem, due to their poor stability, widespread distribution and inefficient gene silencing.
References
W. M. Lee. Hepatitis B virus infection. N. Engl. J. Med. 337(24):1733–1745 (1997).
J. Y. Lau and T. L. Wright. Molecular virology and pathogenesis of hepatitis B. Lancet 342(8883):1335–1340 (1993).
J. R. Wands and H. E. Blum. Primary hepatocellular carcinoma. N. Engl. J. Med. 325(10):729–731 (1991).
D. T. Lau, et al. Long-term follow-up of patients with chronic hepatitis B treated with interferon alpha. Gastroenterology 113(5):1660–1667 (1997).
J. H. Hoofnagle and A. M. di Bisceglie. The treatment of chronic viral hepatitis. N. Engl. J. Med. 336(5):347–356 (1997).
C. L. Lai, et al. A one-year trial of lamivudine for chronic hepatitis B. Asia Hepatitis Lamivudine Study Group. N. Engl. J. Med. 339(2):61–68 (1998).
G. Yao, et al. Long-term effect of lamivudine treatment in chronic hepatitis B virus infection. Zhonghua Gan Zang Bing Za Zhi 7(2):80–83 (1999).
J. R. Wands, et al. Nucleic acid-based antiviral and gene therapy of chronic hepatitis B infection. J. Gastroenterol. Hepatol. 12(9–10):S354–S369 (1997).
F. von Weizsacker, et al. Gene therapy for chronic viral hepatitis: ribozymes, antisense oligonucleotides, and dominant negative mutants. Hepatology 26(2):251–255 (1997).
W. B. Offensperger, et al. Molecular therapeutic strategies in hepatitis B virus infection. Clin. Investig. 72(10):737–741 (1994).
A. Fire, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391(6669):806–811 (1998).
A. P. McCaffrey, et al. Inhibition of hepatitis B virus in mice by RNA interference. Nat. Biotechnol. 21(6):639–644 (2003).
S. B. Kapadia, et al. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc. Natl. Acad. Sci. U. S. A. 100(4):2014–2018 (2003).
J. A. Wilson, et al. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc. Natl. Acad. Sci. U. S. A. 100(5):2783–2788 (2003).
G. A. Coburn and B. R. Cullen. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 76(18):9225–9231 (2002).
I. M. Verma and N. Somia. Gene therapy—promises, problems and prospects. Nature 389(6648):239–242 (1997).
R. I. Mahato, et al. Modulation of gene expression by antisense and antigene oligodeoxynucleotides and small interfering RNA. Expert Opin. Drug Deliv. 2(1):3–28 (2005).
C. Seeger and W. S. Mason. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64(1):51–68 (2000).
D. S. Dane, et al. Virus-like particles in serum of patients with Australia-antigen-associated hepatitis. Lancet 1(7649):695–698 (1970).
H. E. Blum, et al. Hepatitis B virus X protein is not central to the viral life cycle in vitro. J. Virol. 66(2):1223–1227 (1992).
K. Yaginuma, et al. Hepatitis B virus (HBV) particles are produced in a cell culture system by transient expression of transfected HBV DNA. Proc. Natl. Acad. Sci. U. S. A. 84(9):2678–2682 (1987).
T. Shimazu, et al. Post-transcriptional control of the level of mRNA by hepatitis B virus X gene in the transient expression system using human hepatic cells. Genes Cells 3(7):477–484 (1998).
J. Benn and R. J. Schneider. Hepatitis B virus HBx protein activates Ras-GTP complex formation and establishes a Ras, Raf, MAP kinase signaling cascade. Proc. Natl. Acad. Sci. U. S. A. 91(22):10350–10354 (1994).
M. A. Feitelson, et al. Hepatitis B x antigen and p53 are associated in vitro and in liver tissues from patients with primary hepatocellular carcinoma. Oncogene 8(5):1109–1117 (1993).
S. Takada, et al. Disruption of the function of tumor-suppressor gene p53 by the hepatitis B virus X protein and hepatocarcinogenesis. J. Cancer Res. Clin. Oncol. 121(9–10):593–601 (1995).
X. W. Wang, et al. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc. Natl. Acad. Sci. U. S. A. 91(6):2230–2234 (1994).
H. Sirma, et al. Cytosol is the prime compartment of hepatitis B virus X protein where it colocalizes with the proteasome. Oncogene 16(16):2051–2063 (1998).
S. Takada and K. Koike. X protein of hepatitis B virus resembles a serine protease inhibitor. Jpn. J. Cancer Res. 81(12):1191–1194 (1990).
T. H. Lee, et al. Hepatitis B virus X protein interacts with a probable cellular DNA repair protein. J. Virol. 69(2):1107–1114 (1995).
C. Chang, et al. Expression of the precore region of an avian hepatitis B virus is not required for viral replication. J. Virol. 61(10):3322–3325 (1987).
H. S. Chen, et al. The precore gene of the woodchuck hepatitis virus genome is not essential for viral replication in the natural host. J. Virol. 66(9):5682–5684 (1992).
D. R. Milich, et al. The secreted hepatitis B precore antigen can modulate the immune response to the nucleocapsid: a mechanism for persistence. J. Immunol. 160(4):2013–2021 (1998).
D. R. Milich, et al. Is a function of the secreted hepatitis B e antigen to induce immunologic tolerance in utero? Proc. Natl. Acad. Sci. U. S. A. 87(17):6599–6603 (1990).
D. Ganem, et al. Hepatitis B virus reverse transcriptase and its many roles in hepadnaviral genomic replication. Infect. Agents Dis. 3(2–3):85–93 (1994).
J. Hu and C. Seeger. Expression and characterization of hepadnavirus reverse transcriptases. Methods Enzymol. 275:195–208 (1996).
R. Almeida and R. C. Allshire. RNA silencing and genome regulation. Trends Cell Biol. 15(5):251–258 (2005).
Y. Zeng and B. R. Cullen. RNA interference in human cells is restricted to the cytoplasm. RNA 8(7):855–860 (2002).
H. Zhang, et al. Single processing center models for human Dicer and bacterial RNase III. Cell 118(1):57–68 (2004).
G. Meister, et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15(2):185–197 (2004).
G. Tang. siRNA and miRNA: an insight into RISCs. Trends Biochem. Sci. 30(2):106–114 (2005).
A. L. Jackson, et al. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 12(7):1179–1187 (2006).
A. L. Jackson, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA 12(7):1197–1205 (2006).
G. R. Stark, et al. How cells respond to interferons. Annu. Rev. Biochem. 67:227–264 (1998).
H. Akashi, et al. Escape from the interferon response associated with RNA interference using vectors that encode long modified hairpin-RNA. Mol. Biosyst. 1(5–6):382–390 (2005).
T. Watanabe, et al. Intracellular-diced dsRNA has enhanced efficacy for silencing HCV RNA and overcomes variation in the viral genotype. Gene Ther. 13(11):883–892 (2006).
C. A. Sledz, et al. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 5(9):834–839 (2003).
M. Sioud, et al. Suppression of immunostimulatory siRNA-driven innate immune activation by 2′-modified RNAs. Biochem. Biophys. Res. Commun. 361(1):122–126 (2007).
C. R. Allerson, et al. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 48(4):901–904 (2005).
J. A. Hoerter and N. G. Walter. Chemical modification resolves the asymmetry of siRNA strand degradation in human blood serum. RNA 13(11):1887–1893 (2007).
D. A. Braasch, et al. RNA interference in mammalian cells by chemically modified RNA. Biochemistry 42(26):7967–7975 (2003).
D. A. Braasch, et al. Biodistribution of phosphodiester and phosphorothioate siRNA. Bioorg. Med. Chem. Lett. 14(5):1139–1143 (2004).
F. Czauderna, et al. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 31(11):2705–2716 (2003).
J. Soutschek, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 432(7014):173–178 (2004).
A. D. Judge, et al. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Molec. Ther. 13(3):494–505 (2006).
J. Peng, et al. Inhibition of hepatitis B virus replication by various RNAi constructs and their pharmacodynamic properties. J. Gen. Virol. 86(Pt 12):3227–3234 (2005).
D. V. Morrissey, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 23(8):1002–1007 (2005).
T. C. Chu, et al. Aptamer mediated siRNA delivery. Nucleic Acids Res. 34(10):e73 (2006).
C. Wolfrum, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotechnol. 25(10):1149–1157 (2007).
A. M. Derfus, et al. Targeted quantum dot conjugates for siRNA delivery. Bioconjug. Chem. 18(5):1391–1396 (2007).
S. H. Lee, et al. Intracellular siRNA delivery system using polyelectrolyte complex micelles prepared from VEGF siRNA-PEG conjugate and cationic fusogenic peptide. Biochem. Biophys. Res. Commun. 357(2):511–516 (2007).
C. Lorenz, et al. Steroid and lipid conjugates of siRNAs to enhance cellular uptake and gene silencing in liver cells. Bioorg. Med. Chem. Lett. 14(19):4975–4997 (2004).
J. O. McNamara 2nd, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 24(8):1005–1015 (2006).
R. I. Mahato, et al. Cationic lipid-based gene delivery systems: pharmaceutical perspectives. Pharm. Res. 14(7):853–859 (1997).
D. V. Morrissey, et al. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology 41(6):1349–1356 (2005).
R. I. Mahato, et al. Pharmaceutical perspectives of nonviral gene therapy. Adv. Genet. 41:95–156 (1999).
C. Managit, et al. Effect of galactose density on asialoglycoprotein receptor-mediated uptake of galactosylated liposomes. J. Pharm. Sci. 94(10):2266–2275 (2005).
Y. Hattori, et al. Controlled biodistribution of galactosylated liposomes and incorporated probucol in hepatocyte-selective drug targeting. J. Control. Release 69(3):369–377 (2000).
L. A. Sliedregt, et al. Design and synthesis of novel amphiphilic dendritic galactosides for selective targeting of liposomes to the hepatic asialoglycoprotein receptor. J. Med. Chem. 42(4):609–618 (1999).
F. Dasi, et al. Asialofetuin liposome-mediated human alpha1-antitrypsin gene transfer in vivo results in stationary long-term gene expression. J. Mol. Med. 79(4):205–212 (2001).
J. A. Kamps, et al. Massive targeting of liposomes, surface-modified with anionized albumins, to hepatic endothelial cells. Proc. Natl. Acad. Sci. U. S. A. 94(21):11681–11685 (1997).
P. Opanasopit, et al. Inhibition of liver metastasis by targeting of immunomodulators using mannosylated liposome carriers. J. Control. Release 80(1–3):283–294 (2002).
S. Kawakami, et al. Mannose receptor-mediated gene transfer into macrophages using novel mannosylated cationic liposomes. Gene Ther. 7(4):292–299 (2000).
Y. Zhang, et al. Mechanisms of co-modified liver-targeting liposomes as gene delivery carriers based on cellular uptake and antigens inhibition effect. J. Control. Release 117(2):281–290 (2007).
S. Han, et al. Development of biomaterials for gene therapy. Molec. Ther. 2(4):302–317 (2000).
L. M. Santhakumaran, et al. Enhanced cellular uptake of a triplex-forming oligonucleotide by nanoparticle formation in the presence of polypropylenimine dendrimers. Nucleic Acids Res. 32(7):2102–2112 (2004).
A. J. Hollins, et al. Evaluation of generation 2 and 3 poly(propylenimine) dendrimers for the potential cellular delivery of antisense oligonucleotides targeting the epidermal growth factor receptor. Pharm. Res. 21(3), 458–466 (2004).
B. Urban-Klein, et al. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 12(5):461–466 (2005).
M. Grzelinski, et al. RNA interference-mediated gene silencing of pleiotrophin through polyethylenimine-complexed small interfering RNAs in vivo exerts antitumoral effects in glioblastoma xenografts. Hum. Gene Ther. 17(7):751–766 (2006).
S. D. Patil, et al. DNA-based therapeutics and DNA delivery systems: a comprehensive review. AAPS J. 7(1):E61–E77 (2005).
D. A. Wang, et al. Novel branched poly(ethylenimine)-cholesterol water-soluble lipopolymers for gene delivery. Biomacromolecules 3(6):1197–1207 (2002).
W. H. Wen, et al. Targeted inhibition of HBV gene expression by single-chain antibody mediated small interfering RNA delivery. Hepatology 46(1):84–94 (2007).
H. Giladi, et al. Small interfering RNA inhibits hepatitis B virus replication in mice. Molec. Ther. 8(5):769–776 (2003).
C. Klein, et al. Inhibition of hepatitis B virus replication in vivo by nucleoside analogues and siRNA. Gastroenterology 125(1), 9–18 (2003).
O. Milhavet, et al. RNA interference in biology and medicine. Pharmacol. Rev. 55(4):629–648 (2003).
K. Ui-Tei, et al. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 32(3):936–948 (2004).
Y. Naito, et al. siDirect: highly effective, target-specific siRNA design software for mammalian RNA interference. Nucleic Acids Res. 32(Web Server issue):W124–W129 (2004).
B. Yuan, et al. siRNA Selection Server: an automated siRNA oligonucleotide prediction server. Nucleic Acids Res. 32(Web Server issue):W130–W134 (2004).
A. Henschel, et al. DEQOR: a web-based tool for the design and quality control of siRNAs. Nucleic Acids Res. 32(Web Server issue):W113–W120 (2004).
A. D. Judge, et al. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat. Biotechnol. 23(4):457–462 (2005).
D. M. Dykxhoorn, et al. Killing the messenger: short RNAs that silence gene expression. Nat. Rev., Mol. Cell Biol. 4(6):457–467 (2003).
R. M. Schiffelers, et al. Pharmaceutical prospects for RNA interference. Pharm. Res. 21(1):1–7 (2004).
T. Holen, et al. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res. 30(8):1757–1766 (2002).
H. Hohjoh. RNA interference (RNA(i)) induction with various types of synthetic oligonucleotide duplexes in cultured human cells. FEBS Lett. 521(1–3):195–199 (2002).
M. Konishi, et al. Inhibition of HBV replication by siRNA in a stable HBV-producing cell line. Hepatology 38(4):842–850 (2003).
K. Hamasaki, et al. Short interfering RNA-directed inhibition of hepatitis B virus replication. FEBS Lett. 543(1–3):51–54 (2003).
G. Y. Wu and C. H. Wu. Specific inhibition of hepatitis B viral gene expression in vitro by targeted antisense oligonucleotides. J. Biol. Chem. 267(18):12436–12439 (1992).
H. L. Wu, et al. RNA interference-mediated control of hepatitis B virus and emergence of resistant mutant. Gastroenterology 128(3):708–716 (2005).
A. Shlomai and Y. Shaul. Inhibition of hepatitis B virus expression and replication by RNA interference. Hepatology 37(4):764–770 (2003).
S. L. Uprichard, et al. Clearance of hepatitis B virus from the liver of transgenic mice by short hairpin RNAs. Proc. Natl. Acad. Sci. U. S. A. 102(3):773–778 (2005).
C. Ying, et al. Selective inhibition of hepatitis B virus replication by RNA interference. Biochem. Biophys. Res. Commun. 309(2):482–484 (2003).
Y. Chen, et al. Inhibition of hepatitis B virus replication by stably expressed shRNA. Biochem. Biophys. Res. Commun. 311(2):398–404 (2003).
J. Liu, et al. Effect of vector-expressed siRNA on HBV replication in hepatoblastoma cells. World J. Gastroenterol. 10(13):1898–1901 (2004).
M. D. Moore, et al. Stable inhibition of hepatitis B virus proteins by small interfering RNA expressed from viral vectors. J. Gene Med. 7(7):918–925 (2005).
G. L. Ren, et al. Stable inhibition of hepatitis B virus expression and replication by expressed siRNA. Biochem. Biophys. Res. Commun. 335(4):1051–1059 (2005).
T. L. Cheng, et al. Therapeutic inhibition of hepatitis B virus surface antigen expression by RNA interference. Biochem. Biophys. Res. Commun. 336(3):820–830 (2005).
T. R. Brummelkamp, et al. A system for stable expression of short interfering RNAs in mammalian cells. Science 296(5567):550–553 (2002).
P. J. Paddison, et al. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 16(8):948–958 (2002).
P. J. Paddison, et al. Stable suppression of gene expression by RNAi in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 99(3):1443–1448 (2002).
S. M. Elbashir, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411(6836):494–498 (2001).
Q. Ni, et al. Inhibition of human La protein by RNA interference downregulates hepatitis B virus mRNA in 2.2.15 cells. World J. Gastroenterol. 10(14):2050–2054 (2004).
Z. G. Yang, et al. Inhibition of hepatitis B virus surface antigen expression by small hairpin RNA in vitro. World J. Gastroenterol. 11(4):498–502 (2005).
R. S. Tomar, et al. Use of adeno-associated viral vector for delivery of small interfering RNA. Oncogene 22(36):5712–5715 (2003).
L. J. Zhao, et al. Specific gene inhibition by adenovirus-mediated expression of small interfering RNA. Gene 316:137–141 (2003).
E. Devroe and P. A. Silver. Retrovirus-delivered siRNA. BMC Biotechnol. 2:15 (2002).
H. Matta, et al. Use of lentiviral vectors for delivery of small interfering RNA. Cancer Biol. Ther. 2(2):206–210 (2003).
G. Q. Li, et al. Combination of small interfering RNA and lamivudine on inhibition of human B virus replication in HepG2.2.15 cells. World J. Gastroenterol. 13(16):2324–2327 (2007).
S. Carmona, et al. Effective inhibition of HBV replication in vivo by anti-HBx short hairpin RNAs. Molec. Ther. 13(2):411–421 (2006).
C. C. Chen, et al. Long-term inhibition of hepatitis B virus in transgenic mice by double-stranded adeno-associated virus 8-delivered short hairpin RNA. Gene Ther. 14(1):11–19 (2007).
Y. Sun, et al. Effective inhibition of hepatitis B virus replication by small interfering RNAs expressed from human foamy virus vectors. Int. J. Mol. Med. 19(4):705–711 (2007).
S. M. Hammond, et al. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293(5532):1146–1150 (2001).
Z. Chen, et al. Combination of small interfering RNAs mediates greater inhibition of human hepatitis B virus replication and antigen expression. J. Zhejiang Univ. Sci. B 6(4):236–241 (2005).
M. S. Weinberg, et al. Specific inhibition of HBV replication in vitro and in vivo with expressed long hairpin RNA. Molec. Ther. 15(3):534–541 (2007).
D. Jarczak, et al. Hairpin ribozymes in combination with siRNAs against highly conserved hepatitis C virus sequence inhibit RNA replication and protein translation from hepatitis C virus subgenomic replicons. FEBS J. 272(22):5910–5922 (2005).
D. T. Lau, et al. Long-term therapy of chronic hepatitis B with lamivudine. Hepatology 32(4 Pt 1):828–834 (2000).
R. S. Ying, et al. Hepatitis B virus is inhibited by RNA interference in cell culture and in mice. Antivir. Res. 73(1):24–30 (2007).
J. Zhang, et al. Down regulation of viral replication by adenoviral-mediated expression of siRNA against cellular cofactors for hepatitic C virus. Virology, 320(1):135–143 (2004).
Acknowledgements
The project was supported by the National Institutes of Health (R01 DK069968 and R01 DK064633) for R. Mahato and the Department of Public Health of Jiangsu Province of China (H200118) for Y. Chen.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Chen, Y., Cheng, G. & Mahato, R.I. RNAi for Treating Hepatitis B Viral Infection. Pharm Res 25, 72–86 (2008). https://doi.org/10.1007/s11095-007-9504-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11095-007-9504-0