Abstract
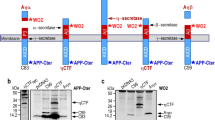
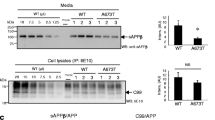
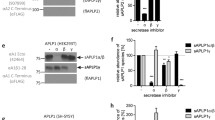
The amyloid peptide (Aβ) deposited in Alzheimer’s disease (AD) is generated by β- and γ-secretase processing of a larger integral membrane protein precursor (APP). Intramembrane processing of APP by γ-secretase also yields an intracellular fragment, CTFγ (a.k.a. AICD), which is highly conserved and is believed to regulate the transcription of several genes including KAI-1 and GSK3β. The intracellular domain of APP is also processed by caspase to a 31 aa fragment that was shown to induce apoptosis by several groups. Although large quantities of CTFγ are generated continuously by neurons, little if any is normally detected in cell lysates, which suggests that it is very rapidly turned over in vivo. Previous studies demonstrated that insulysin (IDE), an Aβ-degrading enzyme, is responsible for cytosol-mediated CTFγ degradation in vitro. Consistent with this finding, knockout mice lacking IDE accumulate CTFγ to detectable levels in the brain, although its levels remain lower than its precursor, suggesting that it continues to be turned over in the brain. Moreover, when we treated cultured cells with IDE inhibitors, we did not observe an increase in CTFγ in cell lysates, suggesting that pathways other than IDE are also involved in CTFγ turnover. To understand CTFγ turnover further, we have mapped the IDE cleavage sites with the intention of mutating them to examine alternative pathways in future studies. Edman degradation revealed that IDE cleaves CTFγ at multiple sites to small peptides ranging from 5 to 14 aa. The cleavage sites do not reveal the existence of any sequence specificity for IDE cleavage. Understanding the turnover mechanisms of CTFγ is critical to the understanding of the signaling function of APP mediated by this fragment. The current study presents the interesting specificity of CTFγ turnover by IDE, which has been previously identified as the major degrading enzyme for Aβ as well as CTFγ. In addition, the study provides evidence for the presence of alternative CTFγ-degrading pathways in the cell.






Similar content being viewed by others
References
Selkoe DJ (2001) Alzheimer’s disease: genes, proteins, and therapy. Physiol Rev 81:741–766
Sambamurti K, Greig NH, Lahiri DK (2002) Advances in the cellular and molecular biology of the beta-amyloid protein in Alzheimer’s disease. Neuromolecular Med 1:1–31
Seubert P, Oltersdorf T, Lee MG, Barbour R, Blomquist C, Davis DL, Bryant K, Fritz LC, Galasko D, Thal LJ et al (1993) Secretion of beta-amyloid precursor protein cleaved at the amino terminus of the beta-amyloid peptide. Nature 361:260–263
Pinnix I, Musunuru U, Tun H, Sridharan A, Golde T, Eckman C, Ziani-Cherif C, Onstead L, Sambamurti K (2001) A novel gamma -secretase assay based on detection of the putative C-terminal fragment-gamma of amyloid beta protein precursor. J Biol Chem 276:481–487
Passer B, Pellegrini L, Russo C, Siegel RM, Lenardo MJ, Schettini G, Bachmann M, Tabaton M, D’Adamio L (2000) Generation of an apoptotic intracellular peptide by gamma-secretase cleavage of Alzheimer’s amyloid beta protein precursor. J Alzheimers Dis 2:289–301
Esch FS, Keim PS, Beattie EC, Blacher RW, Culwell AR, Oltersdorf T, McClure D, Ward PJ (1990) Cleavage of amyloid beta peptide during constitutive processing of its precursor. Science 248:1122–1124
Buxbaum JD, Liu KN, Luo Y, Slack JL, Stocking KL, Peschon JJ, Johnson RS, Castner BJ, Cerretti DP, Black RA (1998) Evidence that tumor necrosis factor alpha converting enzyme is involved in regulated alpha-secretase cleavage of the Alzheimer amyloid protein precursor. J Biol Chem 273:27765–27767
Koike H, Tomioka S, Sorimachi H, Saido TC, Maruyama K, Okuyama A, Fujisawa-Sehara A, Ohno S, Suzuki K, Ishiura S (1999) Membrane-anchored metalloprotease MDC9 has an alpha-secretase activity responsible for processing the amyloid precursor protein. Biochem J 343:371–375
Lammich S, Kojro E, Postina R, Gilbert S, Pfeiffer R, Jasionowski M, Haass C, Fahrenholz F (1999) Constitutive and regulated alpha-secretase cleavage of Alzheimer’s amyloid precursor protein by a disintegrin metalloprotease. Proc Natl Acad Sci U S A 96:3922–3927
Haass C, Selkoe DJ (1993) Cellular processing of beta-amyloid precursor protein and the genesis of amyloid beta-peptide. Cell 75:1039–1042
Chang KA, Suh YH (2005) Pathophysiological roles of amyloidogenic carboxy-terminal fragments of the beta-amyloid precursor protein in Alzheimer’s disease. J Pharmacol Sci 97:461–471
Lu DC, Rabizadeh S, Chandra S, Shayya RF, Ellerby LM, Ye X, Salvesen GS, Koo EH, Bredesen DE (2000) A second cytotoxic proteolytic peptide derived from amyloid beta-protein precursor. Nat Med 6:397–404
Sastre M, Steiner H, Fuchs K, Capell A, Multhaup G, Condron MM, Teplow DB, Haass C (2001) Presenilin-dependent gamma-secretase processing of beta-amyloid precursor protein at a site corresponding to the S3 cleavage of Notch. EMBO Rep 2:835–841
Yu C, Kim SH, Ikeuchi T, Xu H, Gasparini L, Wang R, Sisodia SS (2001) Characterization of a presenilin-mediated amyloid precursor protein carboxyl-terminal fragment gamma. Evidence for distinct mechanisms involved in gamma -secretase processing of the APP and Notch1 transmembrane domains. J Biol Chem 276:43756–43760
Gu Y, Misonou H, Sato T, Dohmae N, Takio K, Ihara Y (2001) Distinct intramembrane cleavage of the beta-amyloid precursor protein family resembling gamma-secretase-like cleavage of Notch. J Biol Chem 276:35235–35238
Weidemann A, Eggert S, Reinhard FB, Vogel M, Paliga K, Baier G, Masters CL, Beyreuther K, Evin G (2002) A novel epsilon-cleavage within the transmembrane domain of the Alzheimer amyloid precursor protein demonstrates homology with Notch processing. Biochemistry 41:2825–2835
Kakuda N, Funamoto S, Yagishita S, Takami M, Osawa S, Dohmae N, Ihara Y (2006) Equimolar production of amyloid beta-protein and amyloid precursor protein intracellular domain from beta-carboxyl-terminal fragment by gamma-secretase. J Biol Chem 281:14776–14786
Cao X, Sudhof TC (2001) A transcriptionally (correction of transcriptively) active complex of APP with Fe65 and histone acetyltransferase Tip60. Science 293:115–120
Kimberly WT, Zheng JB, Guenette SY, Selkoe DJ (2001) The intracellular domain of the beta-amyloid precursor protein is stabilized by Fe65 and translocates to the nucleus in a notch-like manner. J Biol Chem 276:40288–40292
Kim HS, Kim EM, Lee JP, Park CH, Kim S, Seo JH, Chang KA, Yu E, Jeong SJ, Chong YH, Suh YH (2003) C-terminal fragments of amyloid precursor protein exert neurotoxicity by inducing glycogen synthase kinase-3beta expression. Faseb J 17:1951–1953
Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG (2002) Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell 110:55–67
Ryan KA, Pimplikar SW (2005) Activation of GSK-3 and phosphorylation of CRMP2 in transgenic mice expressing APP intracellular domain. J Cell Biol 171:327–335
Cupers P, Orlans I, Craessaerts K, Annaert W, De Strooper B (2001) The amyloid precursor protein (APP)-cytoplasmic fragment generated by gamma-secretase is rapidly degraded but distributes partially in a nuclear fraction of neurones in culture. J Neurochem 78:1168–1178
Cao X, Sudhof TC (2004) Dissection of amyloid-beta precursor protein-dependent transcriptional transactivation. J Biol Chem 279:24601–24611
Mandelkow EM, Drewes G, Biernat J, Gustke N, Van Lint J, Vandenheede JR, Mandelkow E (1992) Glycogen synthase kinase-3 and the Alzheimer-like state of microtubule-associated protein tau. FEBS Lett 314:315–321
Edbauer D, Willem M, Lammich S, Steiner H, Haass C (2002) Insulin-degrading enzyme rapidly removes the beta-amyloid precursor protein intracellular domain (AICD). J Biol Chem 277:13389–13393
Farris W, Mansourian S, Chang Y, Lindsley L, Eckman EA, Frosch MP, Eckman CB, Tanzi RE, Selkoe DJ, Guenette S (2003) Insulin-degrading enzyme regulates the levels of insulin, amyloid beta-protein, and the beta-amyloid precursor protein intracellular domain in vivo. Proc Natl Acad Sci U S A 100:4162–4167
Miller BC, Eckman EA, Sambamurti K, Dobbs N, Chow KM, Eckman CB, Hersh LB, Thiele DL (2003) Amyloid-beta peptide levels in brain are inversely correlated with insulysin activity levels in vivo. Proc Natl Acad Sci U S A 100:6221–6226
Gervais FG, Xu D, Robertson GS, Vaillancourt JP, Zhu Y, Huang J, LeBlanc A, Smith D, Rigby M, Shearman MS, Clarke EE, Zheng H, Van Der Ploeg LH, Ruffolo SC, Thornberry NA, Xanthoudakis S, Zamboni RJ, Roy S, Nicholson DW (1999) Involvement of caspases in proteolytic cleavage of Alzheimer’s amyloid-beta precursor protein and amyloidogenic A beta peptide formation. Cell 97:395–406
LeBlanc A, Liu H, Goodyer C, Bergeron C, Hammond J (1999) Caspase-6 role in apoptosis of human neurons, amyloidogenesis, and Alzheimer’s disease. J Biol Chem 274:23426–23436
Sambamurti K, Refolo LM, Shioi J, Pappolla MA, Robakis NK (1992) The Alzheimer’s amyloid precursor is cleaved intracellularly in the trans-Golgi network or in a post-Golgi compartment. Ann N Y Acad Sci 674:118–128
Li Q, Ali MA, Cohen JI (2006) Insulin degrading enzyme is a cellular receptor mediating varicella-zoster virus infection and cell-to-cell spread. Cell 127:305–316
Brunkan AL, Goate AM (2005) Presenilin function and gamma-secretase activity. J Neurochem 93:769–792
Goate A (2006) Segregation of a missense mutation in the amyloid beta-protein precursor gene with familial Alzheimer’s disease. J Alzheimers Dis 9:341–347
Kopan R, Goate A (2002) Aph-2/Nicastrin: an essential component of gamma-secretase and regulator of Notch signaling and Presenilin localization. Neuron 33:321–324
Pastor P, Goate AM (2004) Molecular genetics of Alzheimer’s disease. Curr Psychiatry Rep 6:125–133
Hecimovic S, Wang J, Dolios G, Martinez M, Wang R, Goate AM (2004) Mutations in APP have independent effects on Abeta and CTFgamma generation. Neurobiol Dis 17:205–218
Bentahir M, Nyabi O, Verhamme J, Tolia A, Horre K, Wiltfang J, Esselmann H, De Strooper B (2006) Presenilin clinical mutations can affect gamma-secretase activity by different mechanisms. J Neurochem 96:732–742
Sambamurti K, Granholm AC, Kindy MS, Bhat NR, Greig NH, Lahiri DK, Mintzer JE (2004) Cholesterol and Alzheimer’s disease: clinical and experimental models suggest interactions of different genetic, dietary and environmental risk factors. Curr Drug Targets 5:517–528
Borg JP, Ooi J, Levy E, Margolis B (1996) The phosphotyrosine interaction domains of X11 and FE65 bind to distinct sites on the YENPTY motif of amyloid precursor protein. Mol Cell Biol 16:6229–6241
Fiore F, Zambrano N, Minopoli G, Donini V, Duilio A, Russo T (1995) The regions of the Fe65 protein homologous to the phosphotyrosine interaction/phosphotyrosine binding domain of Shc bind the intracellular domain of the Alzheimer’s amyloid precursor protein. J Biol Chem 270:30853–30856
Guenette SY, Chen J, Ferland A, Haass C, Capell A, Tanzi RE (1999) hFE65L influences amyloid precursor protein maturation and secretion. J Neurochem 73:985–993
Hoe HS, Tran TS, Matsuoka Y, Howell BW, Rebeck GW (2006) DAB1 and Reelin effects on amyloid precursor protein and ApoE receptor 2 trafficking and processing. J Biol Chem 281:35176–35185
King GD, Scott Turner R (2004) Adaptor protein interactions: modulators of amyloid precursor protein metabolism and Alzheimer’s disease risk? Exp Neurol 185:208–219
Matsuda S, Yasukawa T, Homma Y, Ito Y, Niikura T, Hiraki T, Hirai S, Ohno S, Kita Y, Kawasumi M, Kouyama K, Yamamoto T, Kyriakis JM, Nishimoto I (2001) c-Jun N-terminal kinase (JNK)-interacting protein-1b/islet-brain-1 scaffolds Alzheimer’s amyloid precursor protein with JNK. J Neurosci 21:6597–6607
McLoughlin DM, Miller CC (1996) The intracellular cytoplasmic domain of the Alzheimer’s disease amyloid precursor protein interacts with phosphotyrosine-binding domain proteins in the yeast two-hybrid system. FEBS Lett 397:197–200
Sabo SL, Ikin AF, Buxbaum JD, Greengard P (2003) The amyloid precursor protein and its regulatory protein, FE65, in growth cones and synapses in vitro and in vivo. J Neurosci 23:5407–5415
Sabo SL, Lanier LM, Ikin AF, Khorkova O, Sahasrabudhe S, Greengard P, Buxbaum JD (1999) Regulation of beta-amyloid secretion by FE65, an amyloid protein precursor-binding protein. J Biol Chem 274:7952–7957
Scheinfeld MH, Ghersi E, Laky K, Fowlkes BJ, D’Adamio L (2002) Processing of beta-amyloid precursor-like protein-1 and -2 by gamma-secretase regulates transcription. J Biol Chem 277:44195–44201
Xie Z, Dong Y, Maeda U, Xia W, Tanzi RE (2007) RNA interference silencing of the adaptor molecules ShcC and Fe65 differentially affect amyloid precursor protein processing and Abeta generation. J Biol Chem 282:4318–4325
Zambrano N, Bimonte M, Arbucci S, Gianni D, Russo T, Bazzicalupo P (2002) Feh-1 and apl-1, the Caenorhabditis elegans orthologues of mammalian Fe65 and beta-amyloid precursor protein genes, are involved in the same pathway that controls nematode pharyngeal pumping. J Cell Sci 115:1411–1422
Chen Y, Liu W, McPhie DL, Hassinger L, Neve RL (2003) APP-BP1 mediates APP-induced apoptosis and DNA synthesis and is increased in Alzheimer’s disease brain. J Cell Biol 163:27–33
Leissring MA, Murphy MP, Mead TR, Akbari Y, Sugarman MC, Jannatipour M, Anliker B, Muller U, Saftig P, De Strooper B, Wolfe MS, Golde TE, LaFerla FM (2002) A physiologic signaling role for the gamma -secretase-derived intracellular fragment of APP. Proc Natl Acad Sci U S A 99:4697–4702
Kamal A, Almenar-Queralt A, LeBlanc JF, Roberts EA, Goldstein LS (2001) Kinesin-mediated axonal transport of a membrane compartment containing beta-secretase and presenilin-1 requires APP. Nature 414:643–648
Zheng P, Eastman J, Vande Pol S, Pimplikar SW (1998) PAT1, a microtubule-interacting protein, recognizes the basolateral sorting signal of amyloid precursor protein. Proc Natl Acad Sci USA 95:14745–14750
Shen Y, Joachimiak A, Rosner MR, Tang WJ (2006) Structures of human insulin-degrading enzyme reveal a new substrate recognition mechanism. Nature 443:870–874
Leissring MA, Farris W, Chang AY, Walsh DM, Wu X, Sun X, Frosch MP, Selkoe DJ (2003) Enhanced proteolysis of beta-amyloid in APP transgenic mice prevents plaque formation, secondary pathology, and premature death. Neuron 40:1087–1093
Kurochkin IV (1998) Amyloidogenic determinant as a substrate recognition motif of insulin-degrading enzyme. FEBS Lett 427:153–156
Acknowledgements
We would like to thank the NIH for grants AG028544 and AG023055 supporting this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Special issue in honor of Naren Banik.
Rights and permissions
About this article
Cite this article
Venugopal, C., Pappolla, M.A. & Sambamurti, K. Insulysin Cleaves the APP Cytoplasmic Fragment at Multiple Sites. Neurochem Res 32, 2225–2234 (2007). https://doi.org/10.1007/s11064-007-9449-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11064-007-9449-z