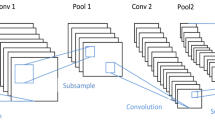
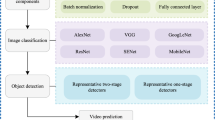
Abstract
Skin diseases have become a challenge in medical diagnosis due to visual similarities. Although melanoma is the best-known type of skin cancer, there are other pathologies that are the cause of many death in recent years. The lack of large datasets is one of the main difficulties to develop a reliable automatic classification system. This paper presents a deep learning framework for skin cancer detection. Transfer learning was applied to five state-of-art convolutional neural networks to create both a plain and a hierarchical (with 2 levels) classifiers that are capable to distinguish between seven types of moles. The HAM10000 dataset, a large collection of dermatoscopic images, were used for experiments, with the help of data augmentation techniques to improve performance. Results demonstrate that the DenseNet201 network is suitable for this task, achieving high classification accuracies and F-measures with lower false negatives. The plain model performed better than the 2-levels model, although the first level, i.e. a binary classification, between nevi and non-nevi yielded the best outcomes.











Similar content being viewed by others
References
American Cancer Society I (ed) (2016) Cancer facts & figures. American Cancer Society, Atlanta
Asha Gnana Priya H, Anitha J, Poonima Jacinth J (2018) Identification of melanoma in dermoscopy images using image processing algorithms. In: 2018 international conference on control, power, communication and computing technologies, ICCPCCT 2018, pp 553–557
Bakheet S (2017) An SVM framework for malignant melanoma detection based on optimized HOG features. Computation 5(1):1–13
Devassy B, Yildirim-Yayilgan S, Hardeberg J (2019) The impact of replacing complex hand-crafted features with standard features for melanoma classification using both hand-crafted and deep features. Adv Intell Syst Comput 868:150–159
Gao Z et al (2019) Privileged modality distillation for vessel border detection in intracoronary imaging. IEEE Trans Med Imaging 39(5):1524–1534
Gao Z, Wang X, Sun S, Wu D, Bai J, Yin Y, Liu X, Zhang H, de Albuquerque VHC (2020) Learning physical properties in complex visual scenes: an intelligent machine for perceiving blood flow dynamics from static CT angiography imaging. Neural Netw 123:82–93
Gao Z, Wu S, Liu Z, Luo J, Zhang H, Gong M, Li S (2019) Learning the implicit strain reconstruction in ultrasound elastography using privileged information. Med Image Anal 58:101534
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
Hussain Z, Gimenez F, Yi D, Rubin D (2017) Differential data augmentation techniques for medical imaging classification tasks. In: AMIA annual symposium proceedings, vol 2017. American Medical Informatics Association, p 979
Jafari MH, Karimi N, Nasr-Esfahani E, Samavi S, Soroushmehr SMR, Ward K, Najarian K (2016) Skin lesion segmentation in clinical images using deep learning. In: 2016 23rd international conference on pattern recognition (ICPR), pp 337–342
Jafari MH, Nasr-Esfahani E, Karimi N, Soroushmehr SMR, Samavi S, Najarian K (2017) Extraction of skin lesions from non-dermoscopic images for surgical excision of melanoma. Int J Comput Assist Radiol Surg 12(6):1021–1030
Jerant AF, Johnson JT, Sheridan C, Caffrey TJ (2000) Early detection and treatment of skin cancer. Am Fam Phys 62(2):357–368, 375–376, 381–382
Khan MA, Javed MY, Sharif M, Saba T, Rehman A (2019) Multi-model deep neural network based features extraction and optimal selection approach for skin lesion classification. In: 2019 international conference on computer and information sciences (ICCIS). IEEE, pp 1–7
Li J, Zhou G, Qiu Y, Wang Y, Zhang Y, Xie S (2019) Deep graph regularized non-negative matrix factorization for multi-view clustering. Neurocomputing 390:108–116
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JA, van Ginneken B, Sánchez CI (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88
Liu N, Wan L, Zhang Y, Zhou T, Huo H, Fang T (2018) Exploiting convolutional neural networks with deeply local description for remote sensing image classification. IEEE Access 6:11215–11228
Mobiny A, Singh A, Van Nguyen H (2019) Risk-aware machine learning classifier for skin lesion diagnosis. J Clin Med 8(8):1241
Moldovan D (2019) Transfer learning based method for two-step skin cancer images classification. In: 2019 E-health and bioengineering conference (EHB), pp 1–4
Nachbar F, Stolz W, Merkle T, Cognetta AB, Vogt T, Landthaler M, Bilek P, B-Falco O, Plewig G (1994) The ABCD rule of dermatoscopy: high prospective value in the diagnosis of doubtful melanocytic skin lesions. J Am Acad Dermatol 30(4):551–559
Nida N, Irtaza A, Javed A, Yousaf M, Mahmood M (2019) Melanoma lesion detection and segmentation using deep region based convolutional neural network and fuzzy C-means clustering. Int J Med Inf 124:37–48
Nugroho AA, Slamet I, Sugiyanto (2019) Skins cancer identification system of HAMl0000 skin cancer dataset using convolutional neural network. AIP Conf Proc 2202(1):020039
Oliveira RB, Papa JP, Pereira AS, Tavares JMR (2018) Computational methods for pigmented skin lesion classification in images: review and future trends. Neural Comput Appl 29(3):613–636
Pai K, Giridharan A (2019) Convolutional neural networks for classifying skin lesions. In: TENCON 2019—2019 IEEE region 10 conference (TENCON). IEEE, pp 1794–1796
Pereira dos Santos F, Antonelli Ponti M (2018) Robust feature spaces from pre-trained deep network layers for skin lesion classification. In: 2018 31st SIBGRAPI conference on graphics, patterns and images (SIBGRAPI). IEEE, pp 189–196
Ruela M, Barata C, Marques J, Rozeira J (2017) A system for the detection of melanomas in dermoscopy images using shape and symmetry features. Comput Methods Biomech Biomed Eng: Imaging Vis 5(2):127–137
Sae-Lim W, Wettayaprasit W, Aiyarak P (2019) Convolutional neural networks using mobileNet for skin lesion classification. In: 2019 16th international joint conference on computer science and software engineering (JCSSE), pp 242–247
Sandler M, Howard A, Zhu M, Zhmoginov A, Chen LC (2018) Mobilenetv2: inverted residuals and linear bottlenecks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4510–4520
Shahin AH, Kamal A, Elattar MA (2018) Deep ensemble learning for skin lesion classification from dermoscopic images. In: 2018 9th Cairo international biomedical engineering conference (CIBEC). IEEE, pp 150–153
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. In: 2015 IEEE conference on computer vision and pattern recognition (CVPR), pp 1–9
Szegedy C, Ioffe S, Vanhoucke V, Alemi AA (2017) Inception-v4, inception-resnet and the impact of residual connections on learning. In: 31st AAAI conference on artificial intelligence
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2818–2826
Thurnhofer-Hemsi K, Domínguez E (2019) Analyzing digital image by deep learning for melanoma diagnosis. In: Proceedings of the 15th international work-conference on artificial neural networks (IWANN), pp 270–279
Tschandl P, Rosendahl C, Kittler H (2018) The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci Data 5:180161
Victor A, Ghalib M (2017) Automatic detection and classification of skin cancer. Int J Intell Eng Syst 10(3):444–451
Yadav V, Kaushik V (2018) Detection of melanoma skin disease by extracting high level features for skin lesions. Int J Adv Intell Paradig 11(3–4):397–408
Yu L, Chen H, Dou Q, Qin J, Heng PA (2017) Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Trans Med Imaging 36(4):994–1004
Zhou T, Thung K, Zhu X, Shen D (2019) Effective feature learning and fusion of multimodality data using stage-wise deep neural network for dementia diagnosis. Hum Brain Mapp 40(3):1001–1016
Acknowledgements
This work is partially supported by the Ministry of Economy and Competitiveness of Spain under Grants TIN2016-75097-P and PPIT.UMA.B1.2017. It is also partially supported by the Ministry of Science, Innovation and Universities of Spain under Grant RTI2018-094645-B-I00, project name Automated detection with low-cost hardware of unusual activities in video sequences. It is also partially supported by the Autonomous Government of Andalusia (Spain) under project UMA18-FEDERJA-084, project name Detection of anomalous behavior agents by deep learning in low-cost video surveillance intelligent systems. All of them include funds from the European Regional Development Fund (ERDF). The authors thankfully acknowledge the computer resources, technical expertise and assistance provided by the SCBI (Supercomputing and Bioinformatics) center of the University of Málaga. They also gratefully acknowledge the support of NVIDIA Corporation with the donation of two Titan X GPUs used for this research. The authors acknowledge the funding from the Universidad de Málaga. Karl Thurnhofer-Hemsi (FPU15/06512) is funded by a PhD scholarship from the Spanish Ministry of Education, Culture and Sport under the FPU program.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Thurnhofer-Hemsi, K., Domínguez, E. A Convolutional Neural Network Framework for Accurate Skin Cancer Detection. Neural Process Lett 53, 3073–3093 (2021). https://doi.org/10.1007/s11063-020-10364-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11063-020-10364-y