Abstract
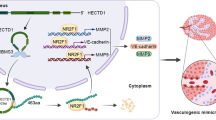
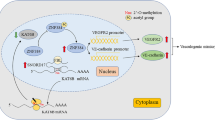
The purpose of this study was to investigate the functions of microRNA-9, which is a tissue-specific microRNA in central nervous system, in the vasculogenic mimicry (VM) of glioma cell lines in vitro and in vivo.Glioma cell lines U87MG, U251 and SHG44 were transfected with microRNA-9 mimic, microRNA-9 inhibitor or scramble sequences. The amount of microRNA-9 and Stathmin (STMN1) mRNA was determined by quantitative real-time PCR, and the protein expression of STMN1 was determined by western blot. Cell proliferation and apoptosis were assessed. The interactions between the 3′UTR of STMN1 and miR-9 was determined by luciferase reporter assay. The VM capacity in vitro was evaluated using VM formation assay, and the rescue experiment of STMN1 was carried out in U251 cells. The in vivo experiment was applied with animal models implanted with U87MG cells.MicroRNA-9 mimic transfection reduced proliferation and increased apoptosis in glioma cell lines (p < 0.05). MicroRNA-9 mimic up-regulated STMN1 mRNA levels but reduced its protein levels (p < 0.05), and luciferase activity of STMN1 was suppressed by microRNA-9 mimic transfection (p < 0.05). Furthermore, microRNA-9 mimic transfection suppressed tumor volume growth, as well as VM both in vitro and in vivo. The cell viability and microtube density were upregulated in U251 cells after STMN1 up-regulation (p < 0.05). STMN1 is a target of microRNA-9, and microRNA-9 could modulate cell proliferation, VM and tumor volume growth through controlling STMN1 expression. MicroRNA-9 and its targets may represent a novel panel of molecules for the development of glioma treatment.





Similar content being viewed by others
References
Rubin CI, Atweh GF (2004) The role of Stathmin in the regulation of the cell cycle. J Cell Biochem 93:242–250
Bhat KM, Setaluri V (2007) Microtubule-associated proteins as targets in cancer chemotherapy. Clin Cancer Res 13:2849–2854
Leibl S, Zigeuner R, Hutterer G et al (2008) EGFR expression in urothelial carcinoma of the upper urinary tract is associated with disease progression and metaplastic morphology. APMIS 116:27–32
Cassimeris L (2002) The oncoprotein 18/Stathmin family of microtubule destabilizers. Curr Opin Cell Biol 14:18–24
Rana S, Maples PB, Senzer N et al (2008) Stathmin 1: a novel therapeutic target for anticancer activity. Expert Rev Anticancer Ther 8:1461–1470
Dong B, Mu L, Qin X et al (2012) Stathmin expression in glioma-derived microvascular endothelial cells: a novel therapeutic target. Oncol Rep 27:714–718
Iorio MV, Croce CM (2009) MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol 27:5848–5856
Omura N, Li CP, Li A, Hong SM et al (2008) Genome-wide profiling of methylated promoters in pancreatic adenocarcinoma. Cancer Biol Ther 7:1146–1156
Bandres E, Agirre X, Bitarte N et al (2009) Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer 125:2737–2743
Hildebrandt MA, Gu J, Lin J et al (2010) Hsa-miR-9 methylation status is associated with cancer development and metastatic recurrence in patients with clear cell renal cell carcinoma. Oncogene 29:5724–5728
Krichevsky AM, King KS, Donachue CP et al (2003) A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 9:1274–1281
Krichevsky AM, Sonntag KC, Isacson O et al (2006) Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells 24:857–864
Delaloy C, Gao FB (2010) A new role for microRNA-9 in human progenitor cells. Cell Cycle 15:2913–2914
Delaloy C, Liu L, Lee JA et al (2010) MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell 4:323–335
Shibata M, Kurokawa D, Nakao H et al (2008) MicroRNA-9 #odulates Cajal-Retzius cell differentiation by suppressing Foxg1 expression in mouse medial pallium. J Neurosci 41:10415–10421
Li Y, Wang F, Lee JA et al (2006) MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Gene Dev 20:2793–2805
Otaegi G, Pollock A, Hong J et al (2011) MicroRNA miR-9 modifies motor neuron columns by a tuning regulation of FoxP1 levels in developing spinal cords. J Neurosci 3:809–818
Bonev B, Pisco A, Papalopulu N (2011) MicroRNA-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Dev Cell 1:19–32
Holmfeldt P, Sellin ME, Gullberg M (2009) Predominant regulators of tubulin monomer-polymer partitioning and their implication for cell polarization. Cell Mol Life Sci 66:3263–3276
Ingber D, Prusty D, Zhengqi S et al (1995) Cell shape, cytoskeletal mechanics and cell cycle control in angiogenesis. J Biomech 28:1471–1484
Petrovic V, Costa RH, Lau LF et al (2008) FoxM1 regulates growth factor-induced expression of kinase-interacting stathmin to promote cell cycle progression. J Biol Chem 283:453–460
Kouzu Y, Uzawa K, Koike H et al (2006) Overexpression of Stathmin in oral squamous-cell carcinoma: correlation with tumour progression and poor prognosis. Br J Cancer 94:717–723
Yuan RH, YM J, Chen HL et al (2006) Stathmin overexpression cooperates with p53 mutation and osteopontin overexpression, and is associated with tumour progression, early recurrence, and poor prognosis in hepatocellular carcinoma. J Pathol 209:549–558
Ghosh R, Gu G, Tillman E et al (2007) Increased expression and differential phosphorylation of stathmin may promote prostate cancer progression. Prostate 67:1038–1052
Wang R, Dong K, Lin F et al (2007) Inhibiting proliferation and enhancing chemosensitivity to taxanes in osteosarcoma cells by RNA interference-mediated down- regulation of Stathmin expression. Mol Med 13:567–575
Alli E, Yang JM, Hait WN (2007) Silencing of Stathmin induces tumor-suppressor function in breast cancer cell lines harboring mutant p53. Oncogene 26:1003–1012
Johnsen JI, Aurelio ON, Kwaja Z et al (2000) p53-mediated negative regulation of Stathmin/Op18 expression is associated with G2/M cell-cycle arrest. Int J Cancer 88:685–691
Ngo TT, Peng T, Liang XJ et al (2007) The 1p-encoded protein stathmin and resistance of malignant gliomas to nitrosoureas. J Natl Cancer Inst 99:639–652
Liang XJ, Choi Y, Sackett DL et al (2008) Nitrosoureas inhibit the stathmin-mediated migration and invasion of malignant glioma cells. Cancer Res 68:5267–5272
Wong QW, Lung EW, Law PT et al (2008) MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin 1. Gastroenterology 135:257–269
Wang J, Gu Z, Ni P et al (2011) NF-kappaB P50/P65 hetero-dimer mediates differential regulation of CD166/ALCAM expression via interaction with micoRNA-9 after serum deprivation, providing evidence for a novel negative auto-regulatory loop. Nucleic Acids Res 15:6440–6455
Myatt S, Wang J, Monteiro L et al (2010) Definition of microRNAs that repression expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer Res 70:367–377
Ma L, Young J, Prabhala H et al (2010) miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Mat cell Biol 3:247–256
Luo H, Zhang H, Zhang Z et al (2009) Down-regulated miR-9 and miR-433 in human gastric carcinoma. J Exp Clin Cancer Res 28:82
Wan H, Guo L, Liu T et al (2010) Regulation of the transcription factor NF-kB1 by micro RNA-9 in human gastric adenocarcinoma. Mole Cancer 9:16
Laios A, O’Toole S, Flavin R et al (2008) Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mole Cancer 7:35
Arora H, Qureshi R, Jin S et al (2011) miR-9 and let-7g enhance the sensitivity to ionizing radiation by suppression of NFkappaB1. Exp Mole Med 5:298–304
Nass D, Rosenwald S, Meiri E et al (2009) MiR-92b and miR-9/9* are specifically expressed in brain primary tumors and can be used to differentiate primary from metastatic brain tumors. Brain Pathol 19:375–383
Jeon H, Sohn Y, Oh S et al (2011) ID4 Imparts chemoresistance and cancer stemness to glioma cells by derepressing miR-9*-mediated suppression of SOX2. Cancer Res 9:3410–3421
Schraivogel D, Weimann L, Beier D et al (2011) CAMTA1 is a novel tumour suppressor regulated by miR-9/9* in glioblastoma stem cells. EMBOJ 20:4309–4322
Ben-Hamo R, Efroni S (2011) Gene expression and network-based analysis reveals a novel role for hsa-miR-9 and drug control over the p38 network in glioblastoma multiforme progression. Genome Med 3(11):77
Chao T, Zhang Y, Yan X et al (2008) Mir-9 regulates the expression of CBX7 in Human Glioma. Zhongguo Yi Xue Ke Xue Yuan Xue Bao 30:268–274
Maniotis AJ, Folberg R, Hess A et al (1999) Vascular channel formation by human melanoma cells in vivo and in vitro: vasculogenic mimicry. Am J Pathol 155:739–752
Folberg R, Maniotis AJ (2004) Vasculogenic mimicry. APMIS 112:508–525
Wang S, Yu L, Ling G et al (2012) Vasculogenic mimicry and its clinical significance in medulloblastoma. Cancer Biol Ther 5:341–348
Chen Y, Jing Z, Luo C et al (2012) Vasculogenic mimicry-potential target for glioma therapy: an in vitro and in vivo study. Med Oncol 29:324–331
Acknowledgments
The authors would like to thank Ms. Shaohong Fang and Mr. Jiangtian Tian form Key Laboratory of Myocardial Ischemia Mechanism and Treatment Ministry, Harbin, China, for technically support.
Conflict of interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Song, Y., Mu, L., Han, X. et al. MicroRNA-9 inhibits vasculogenic mimicry of glioma cell lines by suppressing Stathmin expression. J Neurooncol 115, 381–390 (2013). https://doi.org/10.1007/s11060-013-1245-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-013-1245-9