Abstract
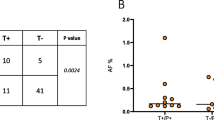
Glioblastoma multiforme (GBM) is the most aggressive brain tumor in adults and remains incurable despite multimodal intensive treatment regimens including surgical resection, radiation and chemotherapy. EGFRvIII is a truncated extracellular mutant of the EGF receptor (EGFR) found in about a third of GBMs. It confers enhanced tumorigenic behavior and is associated with chemo- and radio-resistance. GBM patients testing positive for EGFRvIII have a bleaker prognosis than those who do not. Targeting EGFRvIII positive tumors via vaccines or antibody–drug-conjugates represents a new challenging therapeutic avenue with potential great clinical benefits. In this study, we developed a strategy to detect EGFRvIII deletion in the circulating tumor DNA. The overall goal is to identify a simple and robust biomarker in the peripheral blood of patients diagnosed with GBM in order to follow their disease status while on treatment. Thirteen patients were included in this study, three of which were found to carry the EGFRvIII deletion. The circulating DNA status for EGFRvIII correlates with the analysis performed on the respective tumor samples, and its level seems to correlate with the extent of the tumor resection. This semi-quantitative blood biomarker may represent a strategy to (1) screen patients for an anti-EGFRvIII therapy and (2) monitor the patients’ response to treatment.





Similar content being viewed by others
References
Ohgaki H, Dessen P, Jourde B, Horstmann S, Nishikawa T, Di Patre PL, Burkhard C, Schuler D, Probst-Hensch NM, Maiorka PC, Baeza N, Pisani P, Yonekawa Y, Yasargil MG, Lutolf UM, Kleihues P (2004) Genetic pathways to glioblastoma: a population-based study. Cancer Res 64(19):6892–6899. doi:10.1158/0008-5472.CAN-04-1337
Ohgaki H, Kleihues P (2007) Genetic pathways to primary and secondary glioblastoma. Am J Pathol 170(5):1445–1453. doi:10.2353/ajpath.2007.070011
Ohgaki H, Kleihues P (2005) Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol 64(6):479–489
Burger PC, Green SB (1987) Patient age, histologic features, and length of survival in patients with glioblastoma multiforme. Cancer 59(9):1617–1625
Tait MJ, Petrik V, Loosemore A, Bell BA, Papadopoulos MC (2007) Survival of patients with glioblastoma multiforme has not improved between 1993 and 2004: analysis of 625 cases. Br J Neurosurg 21(5):496–500. doi:10.1080/02688690701449251
Heimberger AB, Hlatky R, Suki D, Yang D, Weinberg J, Gilbert M, Sawaya R, Aldape K (2005) Prognostic effect of epidermal growth factor receptor and EGFRvIII in glioblastoma multiforme patients. Clin Cancer Res 11(4):1462–1466. doi:10.1158/1078-0432.CCR-04-1737
Batra SK, Castelino-Prabhu S, Wikstrand CJ, Zhu X, Humphrey PA, Friedman HS, Bigner DD (1995) Epidermal growth factor ligand-independent, unregulated, cell-transforming potential of a naturally occurring human mutant EGFRvIII gene. Cell Growth Differ 6(10):1251–1259
Beers R, Chowdhury P, Bigner D, Pastan I (2000) Immunotoxins with increased activity against epidermal growth factor receptor vIII-expressing cells produced by antibody phage display. Clin Cancer Res 6(7):2835–2843
Modjtahedi H, Moscatello DK, Box G, Green M, Shotton C, Lamb DJ, Reynolds LJ, Wong AJ, Dean C, Thomas H, Eccles S (2003) Targeting of cells expressing wild-type EGFR and type-III mutant EGFR (EGFRvIII) by anti-EGFR MAb ICR62: a two-pronged attack for tumour therapy. Int J Cancer 105(2):273–280. doi:10.1002/ijc.11055
Perera RM, Narita Y, Furnari FB, Gan HK, Murone C, Ahlkvist M, Luwor RB, Burgess AW, Stockert E, Jungbluth AA, Old LJ, Cavenee WK, Scott AM, Johns TG (2005) Treatment of human tumor xenografts with monoclonal antibody 806 in combination with a prototypical epidermal growth factor receptor-specific antibody generates enhanced antitumor activity. Clin Cancer Res 11(17):6390–6399. doi:10.1158/1078-0432.CCR-04-2653
Fleischhacker M, Schmidt B (2007) Circulating nucleic acids (CNAs) and cancer—a survey. Biochim Biophys Acta 1775(1):181–232. doi:10.1016/j.bbcan.2006.10.001
Goebel G, Zitt M, Muller HM (2005) Circulating nucleic acids in plasma or serum (CNAPS) as prognostic and predictive markers in patients with solid neoplasias. Dis Markers 21(3):105–120
Gormally E, Caboux E, Vineis P, Hainaut P (2007) Circulating free DNA in plasma or serum as biomarker of carcinogenesis: practical aspects and biological significance. Mutat Res 635(2–3):105–117. doi:10.1016/j.mrrev.2006.11.002
Quail MA, Swerdlow H, Turner DJ (2009) Improved protocols for the illumina genome analyzer sequencing system. Curr Protoc Hum Genet Chapter 18:Unit 18 12. doi:10.1002/0471142905.hg1802s62
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10(3):R25
Boeva V, Popova T, Bleakley K, Chiche P, Cappo J, Schleiermacher G, Janoueix-Lerosey I, Delattre O, Barillot E (2012) Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 28(3):423–425
Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29(1):24–26
Frederick L, Eley G, Wang XY, James CD (2000) Analysis of genomic rearrangements associated with EGRFvIII expression suggests involvement of Alu repeat elements. Neuro Oncol 2(3):159–163
Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, Kinzler KW, Vogelstein B, Diaz LA Jr (2008) Circulating mutant DNA to assess tumor dynamics. Nat Med 14(9):985–990
Diehl F, Schmidt K, Durkee KH, Moore KJ, Goodman SN, Shuber AP, Kinzler KW, Vogelstein B (2008) Analysis of mutations in DNA isolated from plasma and stool of colorectal cancer patients. Gastroenterology 135(2):489–498
Taniguchi K, Uchida J, Nishino K, Kumagai T, Okuyama T, Okami J, Higashiyama M, Kodama K, Imamura F, Kato K (2011) Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res 17(24):7808–7815
Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, Antipova A, Lee C, McKernan K, De La Vega FM, Kinzler KW, Vogelstein B, Diaz LA, Jr., Velculescu VE (2010) Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med 2 (20):20ra14. doi:10.1126/scitranslmed.3000702
Yin D, Ogawa S, Kawamata N, Tunici P, Finocchiaro G, Eoli M, Ruckert C, Huynh T, Liu G, Kato M, Sanada M, Jauch A, Dugas M, Black KL, Koeffler HP (2009) High-resolution genomic copy number profiling of glioblastoma multiforme by single nucleotide polymorphism DNA microarray. Mol Cancer Res 7(5):665–677. doi:10.1158/1541-7786.MCR-08-0270
Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321(5897):1807–1812. doi:10.1126/science.1164382
Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, Flanagan A, Teague J, Futreal PA, Stratton MR, Wooster R (2004) The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer 91(2):355–358. doi:10.1038/sj.bjc.6601894
Burke JE, Perisic O, Masson GR, Vadas O, Williams RL (2012) Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110alpha (PIK3CA). Proc Natl Acad Sci U S A 109(38):15259–15264. doi:10.1073/pnas.1205508109
Boisselier B, Gallego Perez-Larraya J, Rossetto M, Labussiere M, Ciccarino P, Marie Y, Delattre JY, Sanson M (2012) Detection of IDH1 mutation in the plasma of patients with glioma. Neurology 79(16):1693–1698. doi:10.1212/WNL.0b013e31826e9b0a
Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky AM, Breakefield XO (2008) Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 10(12):1470–1476. doi:10.1038/ncb1800
Acknowledgments
We would like to thank Dr. Mario Medvedovic and Dr. Jing Chen from the Department of Environmental Health at the University of Cincinnati for their help with the whole genome sequencing data analysis. We would also like to thank Ruth Steele and Susanne Sifri from Neuro-oncology clinical trials unit for their support and Dr. Ady Kendler for pathology. This work was supported by funds from the University of Cincinnati and from the Neuroscience Foundation/Mayfield Clinic.
Conflict of interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Salkeni, M.A., Zarzour, A., Ansay, T.Y. et al. Detection of EGFRvIII mutant DNA in the peripheral blood of brain tumor patients. J Neurooncol 115, 27–35 (2013). https://doi.org/10.1007/s11060-013-1209-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-013-1209-0