Abstract
Although cyclophilin A (CypA) has been reported to be over-expressed in cancer cells and solid tumors, its expression and role in glioblastomas have not been studied. Herein, we show that expression of CypA in human glioblastoma cell lines and tissues is significantly higher than in normal human astrocytes and normal counterparts of brain tissue. To determine the role of over-expressed CypA in glioblastoma, stable RNA interference (RNAi)-mediated knockdown of CypA (CypA KD) was performed in gliobastoma cell line U87vIII (U87MG · ΔEGFR). CypA KD stable single clones decrease proliferation, infiltration, migration, and anchorage-independent growth in vitro and with slower growth in vivo as xenografts in immunodeficient nude mice. We have also observed that knockdown of CypA inhibits expression of interleukin-8 (IL-8), a tumorigenic and proangiogenic cytokine. Conversely, enforced expression of CypA in the CypA KD cell line, Ud-12, markedly enhanced IL-8 transcripts and restored Ud-12 proliferation, suggesting that CypA-mediated IL-8 production provides a growth advantage to glioblastoma cells. CypA knockdown-mediated inhibition of IL-8 is due to reduced activity of NF-κB, which is one of the major transcription factors regulating IL-8 expression. These results not only establish the relevance of CypA to glioblastoma growth in vitro and in vivo, but also suggest that small interfering RNA-based CypA knockdown could be an effective therapeutic approach against glioblastomas.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Malignant gliomas are among the most devastating of cancers, leading to death in most cases [1]. They present unique challenges for therapy due to their difficult access locations, aggressive biological behavior, and diffuse infiltrative growth. Cyclosporin A (CsA) has been reported to have a chemotherapeutic effect in a variety of cancer cells, including human gliomas [2–4]. In a human glioblastoma cell line and glioblastoma cells from brain slices, CsA was shown to affect their growth/survival and migration/invasion [3, 4]. The mechanisms of these effects may relate to the biochemical properties of CsA such as inhibition of cyclophilin isomerase and calcineurin phosphatase activities that mediate immunosuppressive effects that can alter several signal pathways.
CypA is found in normal cells, comprises ~0.6% of total cytosolic protein, and is widely conserved among prokaryotes and eukaryotes [5]. It is a member of the peptidyl-prolyl isomerase (PPI) family, a group of proteins that catalyze cis-trans isomerization of peptidyl-prolyl bonds during protein folding and/or conformational changes [6, 7]. CypA was first identified as the primary intracellular target of the immunosuppressive drug, CsA [8]. The immunosuppressive activity of CsA is thought to result from engagement of calcineurin by the CsA-CypA complex [9], an observation supported by the finding that CypA knockout mice are resistant to immunosuppression by CsA [10].
Several lines of research have revealed that PPIs such as CypA may function as molecular signaling “switches” that can act as novel molecular timers to help control the amplitude and duration of a cellular process [11]. Moreover, the role of CypA nuclear translocation or CypA’s participation in activation of other factors or their nuclear translocation also impacts various cellular functions [12–15]. One recent report demonstrated that knockdown of CypA inhibited signal transducer and activator of transcription 3 (Stat3) interleukin-6-induced tyrosine phosphorylation and nuclear translocation, resulting in altered gene expression in myeloma cell lines [16]. The CypA inhibitor, CsA, exhibited similar effects. Additional data also suggest that CypA may contribute to the pathology of certain human malignancies. Several experiments have demonstrated that CypA is substantially up-regulated in various types of cancers and cancer cell lines [17–20]. These over-expressed CypAs mediate multiple cellular processes that can confer growth advantage to tumor cells in the neoplastic microenvironment. This was substantiated by CypA over-expression in the SILEK-transformed small airway epithelial cell line, which showed that increased CypA expression correlated with dramatically faster tumor formation in vivo [21]. Furthermore, CypA over-expression in cancer cells conferred resistance to anticancer drugs and hypoxia-induced cell death [22, 23]. Therefore, targeted CypA and inhibition of its PPI activity have been suggested as novel therapeutic strategies for treatment of human cancers.
Over-expression of CypA has also been implicated in other pathological processes such as rheumatoid arthritis [24, 25]. In rheumatoid arthritis patients, over-expressed CypA stimulates production of inflammatory cytokines such as tumor necrosis factor alpha (TNF-α), interleukin-1β (IL-1β), IL-8, monocyte chemoattractant protein-1 (MCP-1), and matrix metalloproteinase 9 (MMP-9) [25]. These CypA-stimulated products could arise through a pathway that is dependent on NF-κB activation. However, the means by which CypA mediates signaling to activate NF-κB remains to be investigated. Similarly, stably expressed CypA in the SK-Hep1 cell line revealed that CypA up-regulates the expression of many cytokine-related genes such as IL-8, IL-1β, IL-6, CXCL1, CXCL2, and CXCL3 [22]. These up-regulated cytokines and chemokines may confer tumor cell growth advantage in the neoplastic microenvironment. Therefore, a strategy to reverse these effects might offer attractive options for novel therapeutics.
In human gliomas, IL-8 is expressed and secreted at high levels both in vitro and in vivo, and recent experiments suggest it is critical to glial tumor neovascularity and progression [26]. IL-8 could act as an inflammatory chemoattractant as part of the host response to neoplasia. Moreover, it also acts as a more general proinflammatory factor released in response to tissue stress and necrosis, a proangiogenic factor that promotes new vessel growth, or an autocrine growth factor secreted by tumor cells to promote their own growth. Secretion of IL-8 could also arise from any other cellular compartments such as microglia, macrophages, and neutrophils. IL-8 gene expression and peptide secretion by glioblastoma cells can be dramatically enhanced by TNF-α and IL-1 [27, 28]. The IL-8 promoter contains binding sites for transcription factors NF-κB, AP-1, and C-EBP/NF-IL-6, among others, which are mediated to respond to hypoxia, Fas ligation, death receptor activation, cytosolic calcium, TNF-α, IL-1, other cytokines, and various cellular stresses increase IL-8 expression [29–31].
Expression of CypA and its relationship to highly expressed IL-8 in glioblastoma has not been studied. Herein we report that CypA was strongly expressed in a majority of human glioblastomas and cell lines we tested. This ubiquitous over-expression of CypA in glioblastoma suggests that CypA could be an important component of neoplastic transformation. To determine the role of over-expressed CypA in glioblastoma, stable RNA interference (RNAi)-mediated knockdown of CypA (CypA KD) in a gliobastoma cell line was performed. The CypA KD stable single clones showed decreased proliferation, infiltration, migration, and anchorage-independent growth in vitro, as well as slower tumor growth in vivo. The reversal of the tumor phenotype, and in particular, decreased cell proliferation and slow tumor growth, in CypA KD clones was associated with reduced NF-κB activity and down-regulated IL-8 expression. Thus, targeted knockdown of CypA could be an effective therapeutic approach for glioblastomas.
Materials and methods
Cell culture and cell proliferation assay
Human glioblastoma cell lines U87vIII, U87MG, T98G, Ln229, U138, U378 and immortalized normal human astrocytes were grown in DMEM with low glucose and supplemented with 10% fetal bovine serum. Panc-1 and A549 cells were purchased from ATCC. Panc-1 and A549 cells were cultured in DMEM and RPMI 1640 (Gibco), respectively. Cells were maintained at 37°C in a humidified chamber containing 5% CO2. Cell proliferation was assessed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) method (MTT cell proliferation assay, ATCC). Cells (1500/well) were seeded in triplicate into 96-well plates and cultured for various periods. At each time point, 10 μl of MTT reagent was added to each well and incubated for 2 h, followed by addition of 100 μl of detergent reagent to each well for additional 3 h incubation in the dark. At the end of the reaction, the absorbance in each well was measured at 595 nm in a microtiter plate reader (BIO-TEK). Control wells of medium alone were used as blanks. For the rescue experiments, cells were transfected with vector alone or rescue construct, using lipofectamine 2000 (Invitrogen). Cells were then seeded into 96-well plates 18 h post-transfection. Time zero (0 day) was 6 h after seeding.
Immunohistochemistry staining
A GL802 tissue microarray was purchased from U.S. Biomax, Inc. (Rockville, MD). It contains 35 samples of glioblastoma (grade IV), two samples of adjacent normal tissue, and three samples of normal brain tissue from autopsies, with a duplicate core per sample format. The tissues were formalin-fixed and paraffin-embedded. Array sections were mounted on the positively charged glass slide. Tissue sections were deparaffinized, hydrated, and hybridized with rabbit anti-cyclophilin A antibody (Abcam), followed by incubation of sections with ImmPress anti-rabbit Ig (Vector Lab). After washing the slides three times in buffer 5 min each time, the sections were incubated in peroxidase substrate DAB solution until desired stain intensity developed. Counterstaining with hematoxylin was used for blue coloring of negative nuclei.
Xenograft tumor formation
All animal work described herein was approved by the Chancellor’s Animal Research Committee at UCLA. Following determination of absence of mycoplasma contamination by nucleic acid hybridization, one million cells each of two CypA KD single clones, Ud-12 and Ud-3, scrambled clone (SC), and parental U87vIII were injected in 100 μl DMEM subcutaneously into the flank of 4-week-old, female, NCRU-M immunodeficient mice (Taconic Farms Inc, Hudson, New York); three animals per cell line were used. Tumor dimensions were measured every 2–3 days with calippers and volume was calculated by the formula V = 0.5 × L × W 2, where L is long diameter and W is short diameter.
Reverse transcription and quantitative real-time PCR analysis
Total RNA was isolated from cells, using TRIzol reagent (Invitrogen) according to the manufacturer’s protocols. One microgram of total RNA was used for cDNA synthesis, using Superscript III (Invitrogen) with an oligo(dT)15 primer. Quantitative real-time PCR (QRT-PCR) was performed using the Power SYBR Green PCR Master Mix (Applied Biosystem) and detected by the 7300 Real Time PCR System (Applied Biosystem). The relative expression of IL-8 mRNA normalized to the internal reference 18S rRNA was analyzed using the 2 −ΔΔCT method [32]. The amount of IL-8 transcripts in U87vIII cells, or U87vIII cells transfected with vector alone, was designated as 1.0. Bars indicate the standard error from three individual experiments. The primers used for real-time PCR were: IL-8, 5′-GGGTTGTGGAGAA-GTTTTTG-3′ (forward), 5′-GTTTCACTGGCATCTTCACTG-3′ (reverse); 18S rRNA, 5′-A-GTTGGTGGAGCGATTT-GTC-3′ (forward), 5′-TATTGCTCAATCTCGGGTGG-3′ (reverse).
Scratch-induced migration assay
U87vIII, Ud-3, and scrambled clone were separately seeded onto six-well plates at a density of 5 × 105 per well. After 24 h, confluent monolayers were scratched with a 1 ml plastic pipet tip to create a uniform, cell-free area. Then the cells were incubated at 37°C and photographed at time points 0, 16, and 24 h on an Olympus IX71 inverted microscope using an UPlanFL 4×/0.13 N.A. PhL objective (Olympus). Digital images were acquired with a MicroFire digital camera driven by the PictureFrame imaging software. The distance between the opposing edges was measured using the Adobe PhotoShop software.
Matrigel infiltration assay
An infiltration assay was performed using a six-well plate transwell insert with 8-μm pore size filters (Fisher Scientific) coated with Matrigel (BD Biosciences). Cells (5 × 105) from cell lines U87vIII, Ud-3, Ud-12, and scrambled clone containing 1.5 ml of serum-free DMEM medium, were separately placed into each transwell insert. Each well in the lower chambers was filled with 2.5 ml of DMEM medium containing 10% fetal bovine serum (FBS). After 24 h of incubation at 37°C in a cell culture chamber, the cells on the top surface of the filter were wiped off with cotton swabs. Infiltrated cells on the lower surface of the filter were fixed and stained with crystal violet (Fisher Scientific). Random fields were counted under a light microscope. Olympus microsuiteTM B3SV software was used for photographing.
Anchorage-independent growth in soft agar
To evaluate the ability of individual cell lines to grow in an anchorage-independent manner, 3000 cells from each cell line (U87vIII, Ud-3, Ud-12, and scrambled clone) were each mixed with 0.5 ml of UltraPureTM Agarose (invitrogen) in DMEM medium with 10% FBS (final concentration 0.3%), then overlayed with 1 ml of 0.6% agarose per well of six-well plates (in duplicate). The cells were given 0.5 ml of 10% FBS DMEM per well every 5 days, and incubated for 21 days, then stained by crystal violet. Colonies were counted using a microscope.
IL-8 ELISA
IL-8 ELISA was performed using the Endogen® Human IL-8 ELISA Kit (Thermo Scientific, Inc.) In brief, cells were seeded into 24-well plates 24 h prior to assay, then treated with different chemicals, as described in the Results. Supernatants from cells were collected and analyzed for IL-8 levels according to the manufacturer’s instructions. Absorbance was then measured on an ELISA plate reader set at 450 nm and 550 nm. To correct for optical imperfections on the microplate, 550-nm values were subtracted from 450-nm values. The test samples multiply the interpolated value obtained from the standard curve by a dilution factor to calculate pg/ml of human IL-8 in the sample.
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared using NE-PER nuclear and cytoplasmic extraction reagents (Pierce Biotechnology). EMSA was performed using the Light Shift Chemiluminescent EMSA Kit (Pierce Biotechnology) according to the manufacturer’s directions. In brief, the biotin end-labeled probe (100 fmol) was incubated with nuclear extract (15 μg) and electrophoresed on a native polyacrylamide gel. The DNA was then transferred to a nylon membrane, UV cross-linked, probed with the streptavidin-horseradish peroxidase conjugate, and chemiluminescent substrate was used for detection. The EMSA probes with 5′ biotin labels were purchased from Sigma-Aldrich. Nucleotide sequences were as follows: NF-κB oligo, 5′-AGTTGAGGGGAC-TTTCCCAGGC-3′; AP1 oligo, 5′-CGCTTGATGAGTCAGCCGGAA-3′; C/EBP oligo, 5′-GCCATCAGTTGCAAATCGTGGC-3′. For competition experiments, 100-fold molar excess of unlabeled oligos were incubated with the protein at 25°C for 10 min before addition of labeled probes, and then further incubated for 20 min.
Luciferase reporter assays
The luciferase reporter plasmids driven by −162 to +44 of human IL-8 promoter (p162-IL-8) and its mutants (p162mtAP1, p162mtC/EBP, and p162mtNF-κB) were kindly provided by James J. Pestka (Michigan State University). Cells were plated into 48-well plates and cultured for 1 day, then transfected with 0.3 μg/well of reporter plasmids and 0.006 μg/well Renilla luciferase plasmid, using lipofectamine 2000 (Invitrogen). Firefly and Renilla luciferase activity were measured with the dual-luciferase reporter assay system (Promega). Results are presented as relative reporter activity after normalization to the Renilla luciferase activity. For promoter activity at basal level, cells were assayed 24 h post-transfection. For TNFα and cyclosporin A (CsA) (Sigma-Aldrich) treatment, cells were incubated with 1 μg/ml CsA for 5 h at 24 h post-transfection, then stimulated with 10 ng/ml TNF-α for another 4 h. Cells were then harvested and promoter activity determined.
Statistical analysis
Results are expressed as means ± SE from at least three independent experiments. Statistical analysis was performed using student’s t test. Unless otherwise indicated, P < 0.05 was deemed significant.
Results
Elevated cyclophilin A (CypA) in human glioblastoma cell lines and tissue specimens
CypA is substantially up-regulated in various types of tumors and cancer cell lines [17–21]. To investigate whether elevated CypA at the protein level is also present in glioblastoma cell line, U87MG and its EGFR deletion mutant, U87vIII, and other glioblastoma cell lines, we performed Western blot analysis of cell lysates prepared from these cell lines and compared them to those isolated from normal human astrocytes (NHA). Our data demonstrate that CypA protein expression in glioblastoma cell lines is significantly higher than the control (Fig. 1a, lanes 1–3). This elevated CypA expression is comparable to that seen in the human pancreatic cancer cell line, Panc-1 [17], and the human lung epithelial carcinoma cell line, A549 [21] (Fig. 1a, lanes 4–14). These data clearly demonstrate that CypA is over-expressed in all glioblastoma cell lines we tested.
Over-expression of CypA in glioblastoma cell lines and human tissue specimens. a Elevated CypA at the protein level in glioblastoma cell lines. Five micrograms of cell lysate prepared from U87MG and U87MG. ΔEGFR (U87vIII) and normal human astrocyte (NHA) were subjected to Western blot analysis to detect CypA expression levels (lanes 1–3). Actin was used as a loading control. Over-expression of CypA levels in glioblastoma cell lines is comparable with those in pancreatic and non-small cell lung carcinoma cell lines (lanes 6 and 7). Lanes 8–11 are other glioblastoma cell lines, and U87DK (lane 12) represents K721 mutation and kinase inactive at EGFR. b Relative CypA levels in human glioblastoma and normal tissues. Ten micrograms of tissue lysates were subjected to Western blot analysis. Coomassie brilliant blue R250-stained signals on the nitrocellulose membrane served as internal controls. (N, normal brain tissue; T, tumor tissue). c CypA expression in glioblastoma multiforms (GBMs). Deparaffinized and hydrated tissue sections were hybridized with rabbit anti-cyclophilin A antibody (Abcam), followed by incubating sections with ImmPress anti-rabbit Ig (Vector Lab) and DAB (brown) staining. Arrows indicate typical positive staining for CypA. Non-neoplastic normal cerebrum tissue and cancer-adjacent normal cerebrum tissue were used as controls
To correlate CypA over-expression in glioblastoma cell lines with glioblastoma specimens, eleven human glioblastoma specimens and surrounding normal brain tissue in each specimen were obtain from the laboratory of Dr. Linda Liau (UCLA). Coomassie brilliant blue R250-stained signals on the nitrocellulose membrane served as internal controls prior to immunoblot analysis. As shown in Fig. 1b, the human glioblastoma specimens had 2.1- to 13-fold increases in CypA protein levels compared to adjacent normal tissue. About 73% of tumor tissues examined contained over-expressed CypA, suggesting that CypA could be involved in glioblastoma pathogenesis. In addition, we obtained a GL802 tissue microarray from U.S. Biomax, Inc. (Rockville, MD). Deparaffinized and hydrated tissue sections were hybridized with rabbit anti-cyclophilin A antibody (AbCame), followed by incubation of the sections with ImmPress anti-rabbit Ig (Vector Lab) and DAB (brown) staining. Counterstaining with hematoxylin was used for blue coloring of negative nuclei. CypA immunohistochemical quantitation in glioblastoma multiforms (GBMs) is shown in Table 1. About 86% of tumor tissues contained over-expressed CypA. A representative CypA stain is shown in Fig. 1c.
CypA knockdown correlated with decreased cell proliferation in vitro and slower tumor growth in vivo
Infection of U87vIII cells with the pSilencer-CypA RNAi retrovirus vector yielded five stable single clones with 70–90% CypA knockdown (CypA KD), as quantitated by chemiluminescence of Western blots (Fig. 2a). Among these single clones, Ud-12 had the greatest CypA KD, and the four other single clones (Ud-3, Ud-8, Ud-13 and Ud-15) had lesser amounts. Single clones selected with pSilencer-scrambled RNAi showed no differences in CypA levels relative to parental U87vIII cells (Fig. 2a). To determine whether the CypA KD single clones had an intrinsic effect on cell proliferation, we performed MTT assays to measure cell proliferation in the CypA KD single clones, Ud-3 and Ud-12, and their parental and scrambled control cells. As shown in Fig. 2b, a difference was noted between knockdowns and parental cells and scrambled control cells. Thus, we conclude that the CypA KD single clones Ud-3 and Ud-12 led to decreased proliferation compared to parental U87vIII and SC.
CypA KD single clones result in decreased cell proliferation and tumor growth. a CypA KD stable single clones generated from U87vIII cells. U87vIII cells were seeded into 24-well plates at 5 × 104 cells per well 1 day before infection with enriched viral media prepared from the CypA-specific siRNA insert, scrambled siRNA vector, and linearized retrovirus vector (supplied with the pSilencer H1-Retro kit purchased from Ambion). Cells were transferred into 6-well plates 3 days post-infection and selected with 2 μg/ml puromycin for 2 weeks. Puromycin-resistant clones were harvested and lysed with RIPA buffer supplemented with a protease inhibitor cocktail (Sigma). Five micrograms of total protein was used for Western blot analysis. Membranes probed with actin antibody were used as loading controls. b CypA KD results in decreased cell proliferation. Ud-3 and Ud-12 were compared to parental U87vIII cells and scrambled clone (SC), using MTT assays according to the manufacturer’s protocol. Values of OD 595 absorbance at each time point were the average of triplicate individual readings (bars, SE; *, P < 0.05 compared with parental wild-type cells). c CypA KD decreases tumor growth. Taconic Balb/c/NIH(s) female nu/nu mice were used for xenografts. One million cells were injected. At each time point, tumor dimensions were measured with calipers, and volume was calculated as V= 0.5 × L × W 2. Three mice were used at each time point and each cell type (bars, SE; *, P < 0.05 compared with parental wild-type cells)
To determine whether CypA KD single clones have a similar effect of slowing tumor growth in vivo, we grew CypA KD clones as xenografts in immunodeficient nude mice. CypA KD cells Ud-3 and Ud-12 yielded slower-growing tumors than control cells for the U87vIII cells and SC (Fig. 2c). At 21 days, tumors from CypA KD Ud-12 and Ud-3 cells were, on average, 96 and 74% smaller, respectively, than tumors from control cells. These results indicate that CypA KD had a significant effect on tumor growth in vivo.
CypA KD decreases anchorage-independent growth, cell migration and infiltration in vitro
To determine whether the CypA KD single clones can reverse the transformed phenotype of glioblastoma, we examined anchorage-independent growth of CypA KD, scrambled and parental control cells, using a soft agar assay. After 21 days of incubation, the cells were stained with crystal violet and imaged. Colonies larger than 0.2 mm2 were counted. Results of a typical experiment showed that U87vIII cells and SC formed significant numbers of colonies in soft agar. The Ud-12 clone caused significant decreases in colony numbers (Fig. 3a); notably, Ud-12 also decreased colony sizes. Taken together, these results indicate that CypA KD inhibits anchorage-independent growth of U87vIII cells. Thus, we concluded that CypA KD attenuates the transformed phenotype of U87vIII cells.
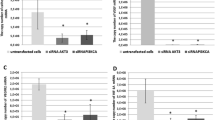
The CypA KD U87vIII single clone decreases anchorage-independent growth, and reduces cell migration and infiltration ability in vitro. a Anchorage-independent growth in vitro. Ud-12 and parental U87vIII cells and SC were plated in duplicate in six-well plates in 0.5% soft agarose on a 1.4% agarose bed agar medium. After 21 days of incubation, the cells were stained with crystal violet and imaged. Colonies larger than 0.2 mm2 were counted. Ud-12/U87vIII= 47.6%, P < 0.05. b Knock down of CypA in U87vIII reduces cell migration. Ud-3 cells were seeded into 6-well plates. When cells reached subconfluent monolayers, a scratch wound was inflicted, and the resulting gap was imaged at the time of the wound and after 16 and 24 h. Images are from a typical experiment. c Infiltration assay in vitro. Transwell plates from Corning Life Sciences were used for invasion assays. The effect of CypA KD on cell infiltration was evaluated using Matrigel as an extracellular matrix component. Chambers were incubated for 24 h under normal cell culture conditions. Invading cells on the lower side of the filter were stained with 0.1% crystal violet (left panel) and quantified after dissolving the cell-bound crystal violet in 10% acetic acid (v/v), followed by measurement of optical density at 540 nm. (Columns, mean of the relative OD 540 nm readings from three independent experiments; bars, SE (right panel); #, P < 0.05 compared with parental wild-type cells)
To investigate the effects of CypA KD on a second manifestation of the transformed phenotype, cell motility of glioblastoma cells, we performed a scratch-induced migration assay. Subconfluent monolayers of CypA KD single clones, parental U87vIII cells, and SC were wounded at time 0 with a plastic scraper to create a cell-free space 2 mm wide, and were monitored at 16 and 24 h after wounding by photomicrograph. The U87vIII cells and SC migrated into the cell-free region, and the wounded area was nearly completely recovered after 24 h (Fig. 3b). However, recovery of the wound area in the CypA KD Ud-3 cells was only 50% at the same time point (Fig. 3b). These results demonstrated that gap closure was strongly attenuated in the CypA KD single clone, Ud-3, which seemed to progress more slowly than the control cells.
Cell infiltration is similar to cell migration, but it requires a cell to migrate through an extracellular matrix or a basement membrane extract barrier, and then become established in a new location. Transwell plates from Corning Life Sciences provide a relatively simple approach to performing cell infiltration assays in vitro. The effect of CypA KD on cell infiltration was evaluated using Matrigel as an extracellular matrix component. Chambers were incubated for 24 h under normal cell culture conditions. Invading cells on the lower side of the filter were stained with 0.1% crystal violet (Fig. 3c) and quantified after dissolving the cell-bound crystal violet in 10% acetic acid (v/v), followed by measurement of optical density at 540 nm (Fig. 3c). Results are presented as the relative OD 540 nm reading.
CypA KD increases apoptosis under the condition of serum withdrawal
It has been reported that over-expression of CypA protects cells from apoptosis [23]. To determine whether CypA KD would sensitize cells to apoptosis under stress conditions, we used a well-established in vitro model, serum withdrawal, to induce apoptosis. The CypA KD single clones, Ud-3 and Ud-12, and parental U87vIII control cells were grown in 96-well plates in media without serum for 6 days. At different time points, cells were subjected to MTT assays to measure cell viability. After 6 days, greater cell death was observed in the KD cells than in the control cells (Fig. 4). As shown in Fig. 4, viable cells of the CypA KD clones, Ud-3 and Ud-12, are significantly reduced on day 6, suggesting that KD cells are more sensitive to serum withdrawal than the control cells.
CypA KD correlates with increased apoptosis under the condition of serum withdrawal. The CypA KD single clones, Ud-3 and Ud-12, and parental U87vIII control cells were grown in 96-well plates in media without serum for 6 days. Cells (10,000/well) were plated in replicates of three. Viable cells were measured at the indicated time points by MTT assays. Columns, mean of the relative OD 595 nm readings from three independent experiments; bars, SE; *, P < 0.05 compared with parental wild-type cells
CypA KD in the U87vIII glioblastoma cell line significantly reduces IL-8 expression
To determine whether CypA KD can affect expression of cytokines such IL-8 in the glioblastoma cell line, U87vIII, real-time RT-PCR was performed to assess cytokine expression in the CypA KD single clones, Ud-12 and Ud-3, as compared to its parental cells and scrambled clone. As shown in Fig. 5a, expression of IL-8 in Ud-12 and Ud-3 cells was significantly decreased by 90 and 80%, respectively, as compared with parental U87vIII control cells. To determine whether down-regulated IL-8 occurs at the protein level, we performed ELISA assays using the Endogen® Human IL-8 ELISA Kit (Thermo Scientifics Inc). As shown in Fig. 5b, the levels of IL-8 expression in the CypA KD clones, Ud-12 and Ud-3, were 98 and 90% lower than parental U87vIII cells. Thus, this observation is very important regarding reversal of tumor phenotype.
CypA KD-mediated down-regulation of IL-8 expression. a IL-8 expression levels were determined by a quantitative real-time PCR analysis. One microgram of total RNA was used for cDNA synthesis, using Superscript III (Invitrogen) with the oligo(dT)15 primer. Quantitative real-time PCR (QRT-PCR) was performed using the Power SYBR Green PCR Master Mix (Applied Biosystem) detected by the 7300 Real Time PCR System (Applied Biosystems). The relative expression of IL-8 mRNA normalized to the internal reference 18S rRNA was analyzed using the 2−ΔΔCT method (columns, mean of results from three independent experiments; bars, SE; *, P < 0.05 compared with parental wild-type cells). b IL-8 ELISA. Cells were seeded into 24-well plates for 24 h. Cell media was collected for measurement of IL-8 expression according the manufacturer’s protocol (columns, mean of results from three independent experiments; bars, SE; *, P < 0.05 compared with parental wild-type cells)
Rescue of the knockdown phenotype restores IL-8 expression and cell proliferation
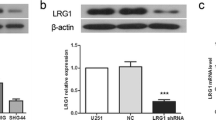
To ensure that the observed knockdown phenotype described above is the result of silencing CypA, the intended target, we introduced siRNA-resistant wild-type CypA with myc-tagged plasmid into Ud-12 cells to reverse siRNA effects. Western blot analysis indicated that exogenous myc-tagged CypA was expressed in Ud-12 cells at levels similar to endogenous CypA in wild-type cells (Fig. 6a). We then analyzed their effects on IL-8 expression and cell proliferation. Results shown in Fig. 6b and c demonstrate that re-introduction of wild-type CypA restored the decreased IL-8 levels and cell proliferation caused by CypA RNAi, indicating that these defects were due to a reduction of CypA. As reported previously, CypA may contribute to proliferation, the inflammatory process, and angiogenesis mediated through activation of cytokine IL-8 [22, 25]. The mechanism for CypA activation of IL-8 and CypA KD reversal remains to be elucidated.
Expression of a siRNA-resistant wild-type CypA rescue expression plasmid restores IL-8 expression and cell proliferation defects. a Western blot analysis of endogenous CypA and exogenous myc-tagged CypA. U87vIII cells (WT) and CypA KD clone Ud-12 were transfected with the pcDNA3.1/Myc.His vector (vector) or the rescue construct (resc_CypA). Five micrograms of each cell extract were subjected to Western blot analysis probing with anti-CypA and anti-myc antibodies. b Real-time PCR analysis of the IL-8 mRNA. The values plotted are the mean of three separate experiments. c MTT assay of cell proliferation. At various time points, cells were subjected to MTT assays. The absorbance in each well was measured at 595 nm in a microtiter plate reader. Bars indicate the standard error from three individual experiments
Identification of CypA-associated transcription factor(s) in regulation of IL-8 expression in U87vIII cells
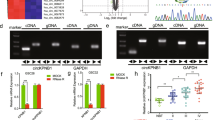
CypA KD inhibits IL-8 expression at both the mRNA and protein levels, suggesting that inhibition of IL-8 expression occurs at the transcriptional level. Although CypA is not a transcription factor and has no DNA-binding motif in the promoter of the IL-8 gene, CypA-associated IL-8 expression may be regulated by transcription factors such as NF-κB, C/EBP, and AP1, which are important in the regulation of IL-8 expression. We used a wild-type and a series of mutant IL-8 promoter luciferase constructs to assess the regulation of IL-8 expression in U87vIII cells mediated by the above transcription factor-binding sites. U87vIII cells were transiently transfected with a −162-bp IL-8 promoter construct and mutation constructs containing AP-1 (−126 to −120), C/EBP (−94 to −81), or NF-κB (−80 to −70) site mutations for 48 h, and then assayed for luciferase activity (Fig. 7a). As shown in Fig. 7b, mutation of the NF-κB, C/EBP, or AP1 sites resulted in a significant decrease of luciferase activity, indicating that all of these sites are necessary for regulation of IL-8 expression in U87vIII cells.
Analysis of IL-8 promoter and regulation of IL-8 expression by TNFα and CsA. a The schematic IL-8 luciferase reporter constructs used in this study. The luciferase reporter plasmids driven by wild-type IL-8 promoter and the mutated promoters containing mutated NF-κB, C/EBP, or AP1 site are designated as wt_IL-8, mt_NF-κB, mt_C/EBP, and mt_AP1, respectively. b U87vIII cells were transfected with indicated reporter plasmids together with Renilla luciferase plasmid. Luciferase activity was measured at 24 h post-transfection (columns, mean of results from three independent experiments; bars, SE). c U87vIII cells were transfected with the reporter plasmid driven by the wild type IL-8 promoter together with Renilla luciferase plasmid. At 24 h post-transfection, cells were incubated with 1 μg/ml CsA or rapamycin for 5 h, then stimulated by 10 ng/ml TNF-α for another 4 h. Cells were then harvested and promoter activity was determined. Results are presented as relative reporter activity after normalization to Renilla luciferase activity. Bars indicate the standard error of three replicates
An attempt to identify CypA-associated transcription factors involved in regulation of the IL-8 promoter in the CypA KD cell line was unsuccessful, due to very low basal levels of IL-8 production. Therefore, we examined whether inhibition of CypA isomerase activity is able to suppress TNFα-induced IL-8 expression in the wild-type U87vIII cells. Cells were treated with CsA, which inhibits CypA isomerase and calcineurin phosphatase activity. Cells were also treated with rapamycin as a control, since rapamycin binds to FKBP-12 but not a calcineurin inhibitor. As shown in Fig. 7C, when the cells transfected with the reporter plasmid driven by the wild type IL-8 promoter were treated with TNFα, luciferase activity increased markedly. This increase was inhibited by CsA but not rapamycin, indicating that CypA isomerase activity is associated with regulation of IL-8 expression in U87vIII cells.
NF-κB activity decreases in CypA KD cells
To further identify the transcription factors responsible for down-regulation of IL-8 expression; we performed EMSAs on nuclear extracts from the CypA KD clones, Ud-3 and Ud-12, to compare to those isolated from parental U87vIII cells and SC. When the NF-κB oligo was used as probe, the density of the protein/DNA complex in CypA KD clones was much lower than in the parental U87vIII cells and SC (Fig. 8a). In contrast, when C/EBP and AP1 oligos were used as probes, no alteration was observed when subjecting these cells to EMSA (Fig. 8b and c). We next examined whether CypA KD affects expression and phosphorylation of IκBα, using Western blot analysis. As shown in Fig. 8d (upper panel), the levels of phosphorylated IκBα are lower in CypA KD clones than in wild-type cells and SC, whereas the levels of IκBα in the wild-type cells and SC are lower than in the CypA KD clones. In Fig. 8d (lower panel), nuclear p65 is lower in CypA KD clones than in wild-type cells and SC, which clearly indicated less nuclear translocated NF-κB in the CypA KD cells and further confirmed our observation above when using EMSA.
Reduced NF-κB binding activity and decreased p65 associated with CypA KD cells. a, b, and c EMSA assays were performed using nuclear extracts from U87vIII cells (WT), scrambled control cells (SC), and the CypA KD clones, Ud-3 and Ud-12, with NF-κB, C/EBP, or AP1 oligos as probes. For competition experiments, protein was pre-incubated with 100-fold molar excess of unlabeled oligos at 25°C for 10 min before addition of labeled probes. p84 expression on Western blots was used as loading control for EMSA. d Cytoplasmic extracts (CE) prepared from U87vIII cells (WT), scrambled control cells (SC), and the CypA KD clones, Ud-3 and Ud-12, were immunoblotted for phosphorylated IκBα (p-IκBα) and IκBα, and actin was used as the internal control (upper panel). Nuclear extracts (NE) were subjected to Western blot analysis probing with anti-p65 antibody. p84 was used as an internal control (lower panel)
Discussion
It has been suggested that CypA may contribute to the pathology of human malignancies. Indeed, several experiments have demonstrated that CypA is substantially up-regulated in various types of cancers and cancer cell lines, including pancreatic adenocarcinoma [17], non-small cell lung cancer [18], hepatocellular carcinoma [33], endometrial carcinoma [19], and esophageal squamous cell carcinoma [20]. These over-expressed CypA molecules mediate multiple cellular processes and confer growth advantage to tumor cells in the neoplastic microenvironment. Further investigation of the connection between CypA and cancer has led to the conclusion that over-expressed CypA is strongly correlated with malignant lung cancer cells [21, 34]. Our data obtained with glioblastoma cell lines also support this concept. A previous report has demonstrated that over-expression of CypA in the SILEK-transformed small airway epithelial cell line correlates with dramatically faster tumor formation in vivo [34], this suggests that the effects of CypA deficiency may have important implications for the treatment of aggressive tumor growth. Our data shows strong evidence that CypA KD leads to decreased cell proliferation in vitro and slower tumor growth in vivo (Fig. 2). Similarly, CypA KD increases apoptosis under the condition of serum withdrawal (Fig. 4), this is a reverse effect for over-expression of CypA in cancer cells renders resistance to anticancer drugs and hypoxia-induced cell death [22, 23]. These results not only establish the relevance of CypA to tumorigenesis, but also substantiate the logic of down-regulating CypA as an approach for treatment of cancer patients.
Over-expression of CypA in rheumatoid arthritis stimulates production of inflammatory cytokines such as TNF-α, IL-1β, IL-8, MCP-1, and MMP-9 [25]. These CypA-stimulated products could occur through a pathway that is dependent on NF-κB activation. However, the mechanism by which CypA mediates signaling to activate NF-κB remains to be investigated. Similarly, stably expressed CypA in the SK-Hep1 cell line revealed that CypA up-regulates the expression of many cytokine-related genes such as IL-8, IL-1β, and IL-6 [22]. These up-regulated cytokines and chemokines may confer tumor cell growth advantage in the neoplastic microenvironment. With these observations in mind, it is conceivable that over-expression of CypA in glioblastomas would lead to highly expressed IL-8. Indeed, in human glioblastoma, IL-8 is expressed and secreted at high levels, and has been suggested to be critical to glial tumor neovascularity and progression [26]. Our data demonstrate that CypA KD in the U87vIII glioblastoma cell line results in profoundly down-regulated IL-8 expression (Fig. 5), suggesting that a strategy to knock down endogenous over-expressed CypA could prevent tumor progression.
IL-8 is known to possess tumorigenic and proangiogenic properties. Down-regulation of IL-8 or its regulatory signaling within the tumor microenvironment would have significant therapeutic potential in modulating tumor progression. A recent report indicated that exposure of U251 glioblastoma cells to calcium inonophore and phorbol-myristate-acetate (PMA) led to dramatic increases in IL-8 expression, which was suppressed by the calcineurin inhibitor, CsA and FK506 [35]. CsA is a CypA inhibitor; therefore, CsA suppression of IL-8 expression may contribute to our observation that CypA KD results in down-regulation of IL-8 expression.
The study conducted in human monocytic leukemia cell line THP-1 demonstrated that CypA induces degradation of IκB and nuclear translocation of NF-κB [25]. Our data concur with their report that CypA KD decreased NF-κB binding activity, decreased phosphorylation and degradation of IκBα, reduced nuclear translocation of NF-κB (Fig. 8d), and led to down-regulation of IL-8 expression. Correspondingly, human recombinant CypA increased phosphorylation of IκBα and stimulated activation of NF-κB in endothelial cells [36]. However, the means by which CypA mediates signaling to activate NF-κB remains to be investigated. A physical and functional map of the human TNF-α/NF-κB signal transduction pathway identified FK506-binding protein 51 (FKBP51), an immunophilin with peptidylprolyl isomerase activity, co-purified with IKKα. This interaction was confirmed by co-immunoprecipitation [37]. The NCBI protein data bank shows that IKKα and IKKβ contain a potential CypA-binding consensus sequence, FRPFLP/FGPXLP. Binding of CypA to IKKα and IKKβ may affect the phosphorylation at Ser 177 and Ser 181 residues that is necessary to activate kinase activity. The possible interaction between CypA and IKKα or IKKβ is currently under investigation.
The present data obtained with both knock down and an ectopically over-expressed rescue plasmid demonstrated that down-regulation and restored IL-8 expression are CypA-dependent (Fig. 6). Although there is no CypA-binding motif in the IL-8 promoter to regulate IL-8 expression, CypA-associated IL-8 production may be regulated by other transcription factors in an indirect manner. Transcriptional regulation of IL-8 has been studied extensively at the level of its promoter region, which contains cis-acting elements for transcription factors C/EBP, AP-1 and NF-κB. Using a wild-type and a series of mutants of IL-8 promoter luciferase constructs, we have found that all three binding sites are important positive regulatory elements for regulation of IL-8. These data suggest that multiple factors are required to work in concert for optimal IL-8 expression in glioblastoma cells. Using EMSA, we found decreased binding of NF-κB to the IL-8 promoter cis-acting element when using nuclear extracts isolated from CypA KD clones (Fig. 8a). These data suggest that CypA is associated with NF-κB binding activity. Taken together, it has been shown that CypA KD leads to decreased NF-κB binding, followed by suppression of a synergistically functional interaction between all three transcription factors resulting in reduced IL-8 expression. Our data demonstrate that knockdown of CypA KD-mediated decreases of cell proliferation, infiltration, and tumor formation may occur through down-regulating IL-8. Thus, targeted knockdown of CypA could be an effective therapeutic approach against devastating glioblastomas.
References
Gurney JG, Kadan-Lottick N (2001) Brain and other central nervous system tumors: rates, trends, and epidemiology. Curr Opin Oncol 13:160–166
Ross HJ, Cho J, Osann K et al (1997) Phase I/II trial of low dose cyclosporine A with EP for advanced non-small cell lung cancer. Lung Cancer 18:189–198
Sliwa M, Markovic D, Gabrusiewicz K et al (2007) The invasion promoting effect of microglia on glioblastoma cells is inhibited by cyclosporine A. Brain 130:476–489
Zupanska A, Dziembowska M, Ellert-Miklaszewska A et al (2005) Cyclosporin A induces growth arrest or programmed cell death of human glioma cells. Neurochem Int 47:430–441
Ryffel B, Woerly G, Greiner B, Haendler B, Mihatsch MJ, Foxwell BM (1991) Distribution of the cyclosporine binding protein cyclophilin in human tissues. Immunology 72:399–404
Fischer G, Aumuller T (2003) Regulation of peptide bond cis/trans isomerization by enzyme catalysis and its implication in physiological processes. Rev Physiol Biochem Pharmacol 148:105–150
Kruse M, Brunke M, Escher A, Szalay AA, Tropschug M, Zimmermann R (1995) Enzyme assembly after de novo synthesis in rabbit reticulocyte lysate involves molecular chaperones and immunophilins. J Biol Chem 270:2588–2594
Handschumacher RE, Harding MW, Rice J, Drugge RJ, Speicher DW (1984) Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science 226:544–547
Bram RJ, Hung DT, Martin PK, Schreiber SL, Crabtree GR (1993) Identification of the immunophilins capable of mediating inhibition of signal transduction by cyclosporine A and FK506: roles of calcineurin binding and cellular location. Mol Cell Biol 13:4760–4769
Colgan J, Asmal M, Yu B, Luban J (2005) Cyclophilin A-deficient mice are resistant to immunosuppression by cyclosporine. J Immunol 174:6030–6038
Lu KP, Finn G, Lee TH, Nicholson LK (2007) Prolyl cis-trans isomerization as a molecular timer. Nat Chem Biol 3:619–629
Colgan J, Asmal M, Neagu M, Yu B, Schneidkraut J, Lee Y, Sokolskaja E, Andreotti A, Luban J (2004) Cyclophilin A regulates TCR signal strength in CD4+ T cells via a praline-directed conformational switch in Itk. Immunity 21:189–201
Song J, Lu Y-C, Yokoyama K, Rossi J, Chiu R (2004) Cyclophilin A is required for retinoic acid-induced neuronal differentiation in p19 cells. J Biol Chem 279:24414–24419
Zhu C, Wang X, Deinum J et al (2007) Cyclophilin A participates in the nuclear translocation of apoptosis-inducing factor in neurons after cerebral hypoxia-ischemis. J Exp Med 204:1741–1748
Pan H, Luo C, Li R et al (2008) Cyclophilin A is required for CXCR4-mediated nuclear export of heterogenous nuclear ribonucleaoprotein A2 activation and nuclear translocation of ERK1/2, and chemotactic cell migration. J Biol Chem 283:623–637
Bauer K, Kretzschmar AK, Cvijic H et al (2009) Cyclophilins contribute to Stat3 signaling and survival of multiple myeloma cells. Oncogene 28:2784–2795
Shen J, Person MD, Zhu J, Abbruzzese JL, Li D (2004) Protein expression profiles in pancreatic adenocarcinoma compared with normal pancreatic tissue and tissue affected by pancreatitis as detected bt two-dimensional gel electrophoresis and mass spectrometry. Cancer Res 64:9018–9026
Campa MJ, Wang MZ, Howard BA, Fitzgerald MC, Patz EF Jr (2003) Protein expression profiling identifies macrophage migration inhibitory factor and cyclophilin A as potential molecular targets in non-small cell lung cancer. Cancer Res 63:1652–1656
Li Z, Zhao X, Bai S, Wang Z, Chen L, Wei Y, Huang C (2008) Proteomics identification of cyclophilin A as a potential prognostic factor and therapeutic target in endometrial carcinoma. Mol Cell Proteomics 7:1810–1823
Qi YJ, He QY, Ma YF et al (2008) Proteomic identification of malignant transformation-related proteins in esophageal squamous cell carcinoma. J Cell Biochem 104:1625–1635
Howard BA, Furumai R, Campa MJ, Rabbani ZN, Vujaskovic Z, Wang XF, Patz EF Jr (2005) Stable RNA interference-mediated suppression of cyclophilin A diminishes non-small-cell lung tumor growth in vivo. Cancer Res 65:8853–8860
Chen S, Zhang M, Ma H, Saiyin H, Shen S, Xi J, Wan B, Yu L (2008) Oligo-microarray analysis reveals the role of cyclophilin A in drug resistance. Cancer Chemother Pharmacol 61:459–469
Choi KJ, Piao YJ, Lim MJ, Kim JH, Ha J, Choe W, Kim SS (2007) Overexpressed cyclophilin A in cancer cells renders resistance to hypoxia- and cisplatin-induced cell death. Cancer Res 67:3654–3662
Arora K, Gwinn WM, Bower MA et al (2005) Extracellular cyclophilins contribute to the regulation of inflammatory responses. J Immunol 175:517–522
Kim H, Kim WJ, Jeon ST, Koh EM, Cha HS, Ahn KS, Lee WH (2005) Cyclophilin A may contribute to the inflammatory processes in rheumatoid arthritis through induction of matrix degrading enzymes and inflammatory cytokines from marcrophages. Clin Immunol 116:217–224
Brat DJ, Bellail AC, Van Meir EG (2005) The role of interleukin-8 and its receptors in gliomagenesis and tumoral angiogenesis. Neuro Oncol 7:122–133
Morita M, Kasahara T, Mukaida N et al (1993) Induction and regulation of IL-8 and MCAF production in human tumor cell lines and brain tumor tissues. Eur Cytokine Netw 4:351–358
Tada M, Suzuki K, Yamakawa Y et al (1993) Human glioblastoma cells produce 77 amino acid interleukin-8. J Neurooncol 16:25–34
Desbaillets I, Diserens AC, de Tribolet N, Hamou MF, Van Meir EG (1999) Regulation of interleukin-8 expression by reduced oxygen pressure in human glioblastoma. Oncogene 18:1447–1456
Hoffmann E, Dittrich-Breiholz O, Holtmann H, Kracht M (2002) Multiple control of interleukin-8 gene expression. J Leukoc Biol 72:847–855
Matsusaki T, Fujikawa K, Nishio Y et al (1993) Taranscription factors NF-IL6 and NF-kappa B synergistically activate transcription of the inflammatory cytokines, interleukin 6 and interleukin 8. Proc Natl Acad Sci USA 90:10193–10197
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408
Lim SO, Park SJ, Kim W, Park SG et al (2002) Proteome analysis of hepatocellular carcinoma. Biochem Biophys Res Commun 291:1031–1037
Howard BA, Zheng Z, Campa MC et al (2004) Translating biomarkers into clinical practice: prognostic implications of cyclophilin A and macrophage migratory inhibitory factor identified from protein expression profiles in non-small cell lung cancer. Lung Cancer 46:313–323
Wakabayashi K, Kambe F, Cao X et al (2004) Inhibitory effects of cyclosporine A on calcium mobilization-dependent interleukin-8 expression and invasion potential of human glioblastoma U251MG cells. Oncogene 23:6924–6932
Jin ZG, Lungu AO, Xie L, Wang M, Wong C, Berk BC (2004) Cyclophilin A is a proinflammatory cytokine that activates endothelial cells. Arterrioscler Thromb Vasc Biol 24:1186–1191
Bouwmeester T, Bauch A, Ruffner H et al (2004) A physical and functional map of the human TNF-α/NF-κB signal transduction pathway. Nat Cell Biol 6:97–105
Acknowledgments
We thank Dr. James J. Pestka (Michigan State University) for IL-8 luciferase reporters, Dr. Kazunari Yokoyama for critical review of the manuscript and discussion, and Rebecca Yang for assistance with experiments.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Sun, S., Wang, Q., Giang, A. et al. Knockdown of CypA inhibits interleukin-8 (IL-8) and IL-8-mediated proliferation and tumor growth of glioblastoma cells through down-regulated NF-κB. J Neurooncol 101, 1–14 (2011). https://doi.org/10.1007/s11060-010-0220-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-010-0220-y