Abstract
We reported that PAX6 suppresses glioblastoma cell growth in vivo and anchorage-independent growth without significant alteration of cell proliferation in vitro, suggesting that PAX6 may alter the tumor microenvironment. Because we found that PAX6 downregulates expression of the gene encoding vascular endothelial growth factor A (VEGFA) in glioma cells, we used a subcutaneous xenograft model to verify PAX6 suppression of VEGFA-induced angiogenesis based on CD31-immunostaining of endothelial cells. The results showed a significant reduction of VEGFA at the transcription level in PAX6-transfected cells in xenografts and PAX6 has a suppressive effect on the microvascular amplification typically seen in glioblastoma. We showed that PAX6 suppression of VEGFA expression requires its DNA binding-domain. The C-terminal truncation mutant of PAX6, however, did not show the dominant negative function in regulating VEGFA expression that it showed previously in regulating MMP2 expression. In the glioma cell line U251HF, we further determined that blocking the PI3K/Akt signaling pathway with either adenoviral-mediated PTEN expression or LY294002 enhanced PAX6-mediated suppression of VEGFA in an additive manner; thus, PAX6-mediated suppression of VEGFA is not via the canonical pathway through HIF1A. These two VEGFA-regulatory pathways can also be similarly modulated in another malignant glioma cell line, U87, but not in LN229 where the basal VEGFA level is low and PTEN is wild-type. PAX6 suppression of VEGFA appears to be blocked in LN229. In conclusion, our data showed that PAX6 can initiate in glioma cells a new signaling pathway independent of PI3K/Akt-HIF1A signaling to suppress VEGFA expression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
PAX6 is an early central-nervous-system (CNS) regulatory gene which has pleotropic effects during development of both the CNS and eye [1–3], and it contributes to both embryonic and adult neurogenesis as a multifunctional player regulating proliferation and differentiation through the control of expression of different downstream molecules in a highly context-dependent manner [4]. We have identified PAX6 as a glioma tumor suppressor gene, based on data showing PAX6 underexpression in higher grade gliomas as opposed to lower grade, in high tumorigenic glioma cell lines in comparison with low- and non-tumorigenic ones, suppression of glioma-cell anchorage-independent growth, invasion, and tumorigenicity, and sensitization of glioma cells to detachment-induced stress [5–8]. Since glioma cells with stable expression of ectopic PAX6 cDNA showed dramatically suppressed growth in vivo, but no significant alteration of cell proliferation in vitro, it appears that PAX6 suppression of glioma tumorigenicity is mediated through changes in the tumor microenvironment, e.g. suppression of tumor angiogenesis.
Microvascular amplification is a histopathology hallmark in diagnosis of glioblastoma multiforme (GBM)—the highest grade and the most common type of glioma [9]. Vascular endothelial growth factor A (VEGFA) is the major angiogenic factor that is overexpressed in GBM [5]. An antiangiogenic agent effective in glioma, Bevacizumab (monoclonal antibody), blocks secreted VEGFA and significantly prolongs survival in patients with malignant glioma. Several small-molecule tyrosine-kinase inhibitors of the VEGFRA receptor show promise as well. Unfortunately despite promising overall results, more then 30% of patients initially fail to respond to anti-angiogenic treatment, and ultimately all patients will relapse and shortly thereafter die of their disease [10]. Understanding pathways abnormally suppressed/activated in glioma cells to affect VEGFA gene expression and VEGFA-induced angiogenesis will improve our understanding of resistance to anti-angiogenic therapies for malignant gliomas.
Enhanced expression of the VEGFA gene by glioma cells was found to occur through the stabilization of hypoxia-inducible factor 1 alpha (HIF1A) under hypoxic conditions [11, 12], through the expression of the gene encoding tissue factor (TF) [13–16], or through direct suppression of promoter activity mediated by the von Hippel-Lindau tumor suppressor gene (VHL) [17]. The action of phosphatase and tensin homolog (PTEN), a tumor suppressor, in suppressing VEGFA expression has been shown to occur via downregulation of HIF1A in a normoxic environment, which is mediated by blocking of the PI3K/Akt signaling pathway—an oncogene pathway active in malignant glioma known to be inactivated by PTEN [18].
By examining gene expressions in four GBMs and their adjacent normal tissues, we found that PAX6 levels were low while VEGFA levels were high in all four of the GBMs relative to adjacent normal tissues from the same patients [19], suggesting that PAX6 may suppress VEGFA gene expression in glioma cells. Here we report our study of PAX6 in glioma cell expression of VEGFA and glioma angiogenesis, and interestingly, co-regulation by PAX6 and PTEN of VEGFA expression in glioma cells.
Materials and methods
Cell lines, stable transfectants, and adenoviral vectors
The following cell lines were described in our previous study [7]: GBM cell lines U251HF, U87 and LN229, and stable PAX6 transfectants of U251HF established through transfection of a plasmid vector (pRC-CMV) expressing 1: wild-type PAX6, 2: C-terminal truncated PAX6 (PAX6-344), 3: mutant form of PAX6 with two mutations (R26G and I87R) in the DNA-binding domain (PD-dmt). The adenoviral vectors for PAX6 (Ad-PAX6) and GFP (Ad-GFP) were described in our previous study [6]. The adenoviral vector for PTEN (Ad-PTEN) and GFP (Ad-GFP) are from Dr. T. J. Liu (M.D. Anderson Cancer Center), which is the same adenovirus type-5 vector as that of Ad-PAX6. Cells were cultured in a DMEM/F12 medium supplemented with or without 5% calf serum in 37°C under normoxic conditions with 5% CO2. A hypoxic condition in experiment was established using a sealed chamber filled with 95% N2 and 5% CO2.
Subcutaneous (s.c.) implantation
Cells of either U251HF or stable transfected U251HF expressing ectopic PAX6 (PAX6_2.2), were suspended in DMEM/F12 medium, and 5 × 106 cell in 50 μl were subcutaneously (s.c.) injected into 4 to 6-week-old female NCrNu-M mice (Taconic, Hudson, NY). Three mice per group were treated, and s.c. injections were anterior to their right and left thighs. Mice were terminated at 25 days after the implantation, and the tumors were dissected, weighted and subjected to RNA extraction as described previously [20]. The s.c. experiment was repeated three times.
Immunohistochemistry analyses of s.c. xenografts
Mice with s.c. xenografts were euthanized, and xenografts were removed and embedded in cryopreservation medium on dry ice. The cryosections were cut 10 μm thick and mounted on cleaned glass slides. After 1 h of 37°C drying, the sections were stored at −80°C. Before immunohistochemistry, sections were removed from −80°C, incubated 5–10 min in the oven at 37°C, and fixed in 4% PFA for 20–30 min at room temperature. Endothelial cells were stained immunohistochemically with primary rat anti-mouse PECAM-1 [CD31] antibody (1:300) from Millipore Corp (Temecula, CA), using hematoxylin to counterstain the nucleus, and detected with biotin-goat anti-mouse SS LINK and DAB detection system from BioGenex (San Ramon, CA). Images were taken on the edge and central part of tumor sections, and blood vessel density (BVD) was calculated. We also performed double-staining of xenograft sections with rat anti-CD31 antibody (1:100) and rabbit anti-PAX6 antibody (1:1,000, kindly provided by Dr. Gary Philips), double-staining with mouse anti-VEGF (1:100) and rabbit anti-PAX6 antibodies, and immunofluorescence detection with fluorescein anti-mouse IgG (H + L) and rhodamine anti-rabbit IgG (H + L) (Minipore Corp), using DAPI to counterstain the nucleus. Image analysis procedure and definitions of microvascular parameters were followed in reference [21].
Modulated imaging (MI)
The MI system has been developed at the Beckman laser institute and medical clinic, and the details of the system have been described previously [22]. We applied the MI technique to determine angiogenesis in vivo non-invasively. Briefly, the MI instrument uses patterned illumination and camera-based detection to obtain quantitative subsurface images of biological tissues over a wide field of view, with information on the optical properties of the tissue being imaged. Imaging at multiple wavelengths (between 650 and 980 nm) provides quantitative measures of the in vivo concentrations of oxy- (OHb), deoxy- (RHb) and total hemoglobin (THb) from subcutaneous xenografts. Oxygen saturation (StO2) is a ratio of OHb to THb.
Real-time absolute quantitative reverse transcription (AqRT-) PCR
The cDNA was reverse transcribed from 0.2 to 2 μg of total RNA from the glioma cell lines, transfectants, and s.c. xenografts using 5 units of Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA). The cDNA was diluted 30 times with 10 mM Tris–HCl (pH = 7.5) and quantified for VEGFA, VHL, and internal reference genes (GAPDH and ENO1) by real-time PCR with human-gene-specific primers (Ziren Research, Irvine, CA). Standard curves were used to determine quantities of the transcripts in cDNA samples. Absolute quantification of VEGFA and VHL expressions relative to reference genes (GAPDH or ENO1) was achieved by using the single standard for both target and reference genes provided by Ziren Research LLC (Irvine, CA). The primer set for VEGFA was designed to amplify all seven transcription variants of VEGFA reported in GeneBank. The primer set for GAPDH and ENO1 were designed to avoid amplification of pseudogenes that have high sequence similarities to the genes’ cDNA sequences. The primer set for VHL specifically amplifies VHL transcription variant 1 (NM_000551). Primer sequence information is available upon request to Ziren Research LLC (www.zirenresearch.com).
Western hybridization and VEGFA enzyme immunometric assay
Whole cell lysate extraction, western blotting, and PAX6 antibody were described previously [17]. Antibodies of Akt, Phopho-Akt (Ser473), and Phospho-GSK3b (Ser9) were from Cell Signaling Technology (Beverly, MA), while PTEN (A2B1) was from Santa Cruzis (Santa Cruz, California), and Actin was from EMD Bioscience (San Diego, CA). A human VEGFA enzyme immunometric assay (ELISA) kit (Assay Designs, Ann Arbor, MI) was used for quantification of the secretory VEGF165 in the conditioned media according to the manufacturer’s instructions.
VEGFA promoter luciferase assay
The VEGFA promoter-luciferase constructs were a generous gift from V. P. Sukhatme (Beth Israel Deaconess Medical Center, Harvard Medical School) [17]. Transient transfection assay were performed using cells (250,000 cells per 35 mm dish plated 1 day before transfection) of U251HF and three clones of U251HF expressing ectopic PAX6 with 0.1 μg pRL-CMV expressing Renilla luciferase as an internal control together with 1 μg VEGFA promoter constructs. Cell were cultured in serum-free medium for 48 h before harvesting cells for detection of luciferase activity using a Dual-Luciferase Reporter Assay System (Promega, Madison, WI) and a TD-20/20 Luminometer (Turner Designs, Sunnyvale, CA).
Infection of Ad-PAX6 and Ad-PTEN in GBM cells
Cells (U251HF, U87 and LN229) in triplicate were plated in 35-mm wells at a density of 4 × 105 cells per well and cultured overnight in DMEM/F12 medium supplemented with 5% calf serum. Cells were then infected in serum-free medium with adenovirus, 50 viral particles per cell for single infection and 25 viral particles per cell for double infection of Ad-PAX6 and Ad-PTEN together or 1 day apart. Cells were grown in normoxic or hypoxic (95% N2, 5% CO2) for 24 or 48 h after infection, and then subjected to RNA extraction and cDNA synthesis for real-time AqRT-PCR. The medium was taken for VEGFA ELISA.
Statistical analysis
Data from real-time AqRT-PCR, ELISA, and luciferase assays were log-transformed to stabilize variance and reduce right-skewing. In some experiments, data were further normalized to the mean of the control treatment within each independent experiment replication. Depending on the design and number of groups, statistical analysis consisted of two-sample t-test, one-way ANOVA or randomized-block ANOVA. Multiple-comparison adjustments in ANOVA post-hoc analysis included Dunnett’s procedure for pairwise comparisons of treatment groups to the control group, and Bonferroni’s procedure for other pre-planned comparisons of interest. All analyses were conducted using SAS version 9.1.3 (The SAS Institute, Cary, NC).
Results
PAX6 suppress angiogenesis in glioma subcutaneous xenografts
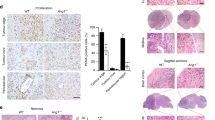
We have tested multiple clones of the stable PAX6-transfected glioblastoma cell line U251HF in our previous study, and concluded that it was PAX6, not the clonal artifact, that suppressed U251HF cells’ anchorage-independent growth, tumorigenicity and invasiveness, and that the vector-transfected clones showed no significant difference in these functional aspects to parental cells [6, 7]. Here we investigated PAX6 regulation of glioma-cell-induced angiogenesis in subcutaneous (s.c.) xenografts derived from one of the transfectants (PAX6_2.2), in comparison with that from the parental cells. Consistent with our previous finding, the PAX6_2.2 cells form significantly smaller tumors compared to the parent cell U251HF after s.c. implantation in nude mice (Fig. 1a). Immunohistochemical (IHC) analysis of endothelial cell marker CD31, as shown in Fig. 1b, reveals that there is a significant reduction in the number and size of blood vessels in xenografts of PAX6_2.2 compared to that of U251HF. IHC analysis revealed severe central necrosis in U251HF s.c. xenografts 25 days after implantation, and significantly fewer blood vessels in the central necrotic area compared to the periphery. We thus compared the blood vessel density (BVD) in tumor periphery regions of U251HF xenograft to that from an overall region of xenografts from PAX6-2.2 that showed a more even distribution of blood vessels. As shown in Fig. 1c, there is a significantly lower BVD in tumor from PAX6_2.2 cells (Fig. 1c).
PAX6 overexpression in glioma cell line U251HF suppresses cell tumorigenicity, tumor angiogenesis and VEGFA expression. a Comparison of the weights of U251HF (n = 8) and its PAX6 stable transfectant PAX6-2.2 xenografts (n = 24) 24 days after subcutaneously implantation in nude mice. Fold change is 3.85; P < 0.0001 by two-sample t-test. b Immunofluroscent analysis of the xenografts from a for expression of PAX6 and CD31, with counterstaining of nuclei by DAPI. See [21] for image analysis procedure and definitions of microvascular parameters. c Comparison of blood vessel density (BVD) normalized to peripheral region of U251HF tumor. d Comparison of total hemoglobin (THb) concentration and tissue oxygen saturation (StO2) in the two groups of s.c. xenografts (four tumors each group, two mice per group) by modulated imaging, all were subjected to CD31-IHC for BVD data above. Two-sample t-test P values were 0.043 for THb, and 0.70 for StO2, after Tukey-adjusted for comparisons among groups. e VEGFA expression in the s.c. xenografts from a by real-time AqRT-PCR. Fold change is 2.52 as shown; P = 0.025 by two-sample t-test
To confirm histological data, we analyzed a group of mice carrying the tumor with modulated imaging (MI) to monitor the blood uptake and consumption in tumor in vivo [22]. MI data revealed that PAX6-2.2 xenografts have significantly lower levels of both oxy-hemoglobin (OHb) and deoxy-hemoglobin (RHb) compared to U251HF xenograft. Consequently, as shown in Fig. 1d, PAX6-2.2 xenografts have a significantly lower level of total hemoglobin (THb) (sum of OHb and RHb), but no significant difference in oxygen saturation (StO2) (ratio of OHb to THb). Therefore, increasing PAX6 level in U251HF cells does not change cellular metabolic activity, but does suppress tumor angiogenesis.
PAX6 suppresses VEGFA expression in vivo
We compared the VEGFA gene expression in the glioma cells in s.c. xenografts shown in Fig. 1, based on data from real-time AqRT-PCR using human-specific gene primers unable to amplify the murine homologues. As shown in Fig. 1e, we found a significantly lower level of VEGFA expression in the s.c. xenografts of PAX6_2.2 compared to those of U251HF; thus, PAX6-suppression of VEGFA takes place in vivo as well as in vitro.
We then quantified the VEGFA expression in U251HF and PAX6_2.2 s.c. xenografts in a different experiment in which we remove the tumor at different times after implantation. As respectively shown in Fig. 2a and b, normalized VEGFA expression was found always higher, and PAX6 always lower, in U251HF xenografts compared to PAX6-2.2 xenografts that were grown for the same period of time in vivo. However, PAX6 in the PAX6-2.2 xenografts showed a 20% decrease in 15-day tumors compared to 11-day tumors (Fig. 2b), suggesting a negative selection pressure on the cells overexpressing PAX6 in vivo. Consistent with this finding, immunostaining of PAX6 in the 24-day PAX6_2.2 tumors showed loss of PAX6 signal in some tumor cells (Fig. 2d). Co-staining for PAX6 and VEGFA in PAX6_2.2 tumors showed distinct areas of cells with expression of either PAX6 or VEGFA but not both. This is consistent with the trends in Fig. 2a and b, which show that the decrease in PAX6 correlates with the increase in VEGFA in overall tumor mass. Importantly, the transient changes in PAX6 and VEGFA mRNA levels in the tumors are related to increase of the tumor volumes (Fig. 2c). Overall, these results indicate that PAX6-mediated suppression of VEGFA expression is responsible for the suppression of tumorigenicity of gliomas, which is shown to be related with suppression of angiogenesis.
Decrease of PAX6 is related to increase of VEGFA expression during tumor growth. a–b Comparison of PAX6 and VEGFA mRNA levels in U251HF and PAX6-2.2 cells grown in vitro or in vivo under nude mice skin at various time. The gene expression was quantified by real-time qRT-PCR, normalized to GAPDH, and compared with U251HF arbitrarily set to unity. c Comparison of tumor volumes that were measured 6 days after s.c. implantation of U251HF and PAX6-2.2. d Co-immunostaining PAX6_2.2 xenografts 24 days after implantation showing single pictures of PAX6 staining and VEGFA staining and merge picture of PAX6 and VEGFA, with cell nucleus counterstained by DAPI
PAX6 downregulation of VEGFA expression at transcription level requiring its DNA-binding domain
We then examined in vitro cell cultures under normoxic condition for changes on secretion of VEGFA in conditioned medium by U251HF and multiple clones of its stable transfectants of wild-type or mutant forms of PAX6. As shown in Fig. 3a, it is the expression of wild-type PAX6, not the mutant forms of PAX6, that changed the VEGFA level secreted by the modified cells.
PAX6 regulation of VEGFA expression. a Comparison of secreted VEGFA quantified by VEGF-165 ELISA from condition medium of cells expressing transfected cDNA constructs of wild-type or two mutant forms of PAX6 (dmt and 344) to the parental glioma cell U251HF, which was arbitrarily set to unity. Bonferroni adjusted P-value with fold of increase (+) and decrease (−) were shown with averages for the clones use for comparison. On top depicts the PAX6 mutants in functional domains, with PAI and RED and HD represent three DNA-binding domain of PAX6, with two point mutations in the paired domain R26G and I87R reported to cause loose of in vitro DNA binding ability [7]. b Luciferase assays of VEGF promoter activities in U251HF and three of U251HF PAX6 transfected clones, with comparison of −52/+144 VEGF promoter activities between parental and PAX6-transfected cells. Mean and SD were from 4 to 8 repeats of transfection and luciferase assay, which were carried out in 2–4 independent experiments
We further investigated PAX6 regulation of VEGFA promoter activity. The transcriptional regulation of the VEGFA promoter seems to play the pre-eminent role in the control of VEGFA expression, and several response elements within the VEGFA promoter region have been characterized in vitro. A key region of the VEGFA promoter, located at approximately −930 from the transcription initiation site, is a target for a number of transcription factors, including HIF1A, TP53, VHL, and SP1. We thus investigated VEGFA promoter activity by transfection of luciferase constructs containing various regions of the VEGFA promoter (2.6, 1.5, 0.35 and 0.2 kb as described previously [17]) in U251HF and its PAX6-transfectants. Since the suppressive effects of PAX6 on the VEGFA promoter were observed in all promoter constructs in PAX6_2.2 cells (data not shown), we focused on examining the 0.2 kb VEGFA promoter, which contains the −52/+144 promoter sequence. This region does not contain any of the binding sites for HIF1A, TP53, VHL, and SP1. As shown in Fig. 3b, the −52/+144 VEGFA promoter activity was lower in all three PAX6-transfectants compared to the parental cell line U251HF; the average of activities relative to U251HF among the three transfectants was 40% (P < 0.0001). Overall, the data showed that PAX6 suppressed VEGFA expression in glioma cells under normoxic in vitro and subcutaneous in vivo conditions.
PAX6-mediated regulation of VEGFA expression is independent of HIF1A or VHL
Although VEGFA expression is regulated via complex pathways, regulation of HIF1A synthesis and stability has been considered to be the central mechanism underlying regulation of VEGFA expression. The tumor suppressor PTEN was reported to suppress VEGFA expression by blocking the PI3K/Akt signaling pathway with subsequent reduction of HIF1A protein level under a normoxic environment [18]. We thus examined the possible influence of PAX6 on regulation of VEGFA through the PI3K/Akt signaling pathway. Western blots were used to detect the activation of the PI3K/Akt signaling pathway in response to adenoviral-mediated overexpression of PAX6 in glioma cells. The result showed that PAX6 did not attenuate the phosphorylation of Akt and GSK3B (Fig. 4a), nor the level of HIF1A (Fig. 4b), in contrast to what was found with adenoviral-mediated overexpression of PTEN in glioma cells.
PAX6 does not alter PI3K/Akt signaling and HIF1A level in glioma cells under normoxic condition. By Western blot assay, phosphorylation status of Akt and the downstream target GSK-3β (a) and HIF1A level (b) were shown unchanged in U251HF cells 48 h after treatment of cells with of Ad-PAX6. Analysis of cells with Ad-PTEN infection was positive control for antibodies, and detections of Actin as control equal protein loading while PAX6 and PTEN for positive infections. c AqRT-PCR revealed the level of VHL expression relative to ENO1 in U251HF cells with or without adenoviral infections. The mean and SD are based on data of two independent experiments, with viral dose of 10 viral partial per cell, cultured for 48 h under serum-free condition after 1 h of infection. P-values are shown for comparisons to mock infection, and are computed using Dunnett’s ANOVA post-hoc procedure
Data from real-time AqRT-PCR quantification of the VHL gene expression revealed a high expression of VHL in U251HF cells: more than 4-fold higher than the level of the reference gene ENO1 (Fig. 4c, first column). We further examined VHL expression 48 h following infection of U251HF cells with adenovirus of PAX6 (Ad-PAX6), with Ad-PTEN, or with both, in two independent experiments. Compared to the mock infection, Ad-PAX6 transfection produced a reduction in the VHL/ENO1 expression ratio of 25% when used alone (Fig. 4c, second column; P = 0.035) and 27% when co-transfecting with Ad-PTEN (Fig. 4c, fourth column; P = 0.026), whereas transfection with Ad-PTEN alone produced a 36% increase in VHL/ENO1 (Fig. 4c, third column; P = 0.027). Since the VHL-expression changes in U251HF cells did not support a suppressive function on VEGFA expression following overexpression of PAX6 alone or together with PTEN, it implies that VHL is unlikely to contribute to PAX6 suppression of VEGFA expression.
PAX6 suppression of VEGFA expression was enhanced by blocking PI3K/Akt signaling pathway
Based on data shown above, PAX6 does not activate the PI3K/Akt signaling pathway, which suggests that the mechanism by which PAX6 suppresses VEGFA differs from that by which PTEN suppresses VEGFA. In agreement with this conclusion, we observed that co-transfection of U251HF cells with Ad-PAX6 and Ad-PTEN enhanced suppression of VEGFA at the levels of both mRNA (Fig. 5a) and secreted VEGFA (Fig. 5b), while no change was seen from transfection with Ad-GFP, excluding possible viral effect. Statistical analysis showed that PAX6 and PTEN work on mRNA levels in an additive manner, i.e. with no significant synergism or antagonism (Table 1). The significant main effects coupled with the lack of significant interaction shows that co-transfection with PAX6 and PTEN produces significantly more suppression of VEGFA expression than either mono-transfection.
PAX6 suppression of VEGFA expression was enhanced by co-expression of PTEN or blocking Akt-signaling pathway in U251HF cells under normoxic condition. Comparison of VEGFA expression in U251HF cells after treatment Ad-PAX6 and/or Ad-PTEN, and/or PI3K inhibitor LY294002 at mRNA level (a) and at secreted protein level (b). Bonferroni adjusted P value were shown
Consistent with PTEN blockage of PI3K/Akt signaling to mediate suppression of VEGFA expression, treatment of cells with the PI3K inhibitor, LY294002, also gave rise to reduction of VEGFA expression in a dose-dependent manner. Co-treatment of Ad-PAX6 and LY294002 further increased the suppression of VEGFA. As shown in Fig. 5a, treatment of U251HF cells with LY294002 alone for 48 h decreased VEGFA expression, to 72% (normalized to GAPDH) and 64% (normalized to ENO1) with a 2 μM dose, and to 65% (normalized to GAPDH) and 49% (normalized to ENO1) with a 10 μM dose. In the same culture condition, infection of Ad-PAX6 alone at a lower dose (10 viral particles per cell) decreased the expression of VEGFA down to 74% (normalized to GAPDH) and 81% (normalized to ENO1). However, combined treatment with Ad-PAX6 and LY294002 enhanced the overall suppression of VEGFA, yielding expression levels of 55% (normalized to GAPDH) or 53% (normalized to ENO1) with 2 μM LY294002, and expression levels of 44% (normalized to GAPDH) or 37% (normalized to ENO1) with 10 μM LY294002. Statistical analysis showed that PAX6 and LY294002 work in an additive manner, i.e. with no significant synergism or antagonism (Table 1).
Similar effects on the enhanced suppression of VEGFA by co-overexpression of PAX6 and PTEN were observed in another glioma cell line, U87. Interestingly, such enhancement can be achieved only when the cells were co-infected by Ad-PAX6 and Ad-PTEN, regardless of whether conditions were normoxic or hypoxic (Fig. 6). Changes in cell viability after vial infection were determined to be minimal based on a Trypan Blue exclusion assay.
Regulation of VEGFA expression in U87 and LN229 by PAX6 or blocking of PI3K/Akt signaling. a VEGF-165 ELISA data on the secreted VEGFA protein in condition medium of cells infected with Ad-PAX6 alone or together with Ad-PTEN simultaneously or separate with 1 day apart, under normoxic and hypoxic conditions. Mean and SD were from 1 to 2 independent experiments, with P-values shown for comparison to mock infection. b Comparison of VEGFA expression in LN229 cells after treatment Ad-PAX6 and/or Ad-PTEN, and/or PI3K inhibitor LY294002 at mRNA level. c comparison of PAX6 and VEGFA expressions in U251HF, U87 and LN229. X marks zero expression value for PAX6 in LN229; PTEN status (mt, mutant or wt, wild type) in these cell lines is based on information in [23]
Infection of another glioma cell line LN229 with Ad-PAX6 alone or together with Ad-PTEN at a lower dose (10 viral particles per cell) decreased the expression of VEGFA down to 70% (normalized to GAPDH or ENO1) (Fig. 6b). But since Ad-GFP also caused a similar level of reduction when normalized to GAPDH, it brings the attention to viral effect in LN229. Trypan Blue exclusion assay showed that LN229 is sensitive to adenoviral infection as well as LY294002 treatment, which apparently cause reductions of ENO1 expression and increase of VEGFA/ENO1 values in Ad-GFP and LY294002 treatments. In contrast to Ad-PAX6, Ad-PTEN slightly increased VEGFA expression in LN229 by about 1.5 fold (Fig. 6b). Glioma cell line LN229 is one of the few glioma cell lines that has a wild-type form of the PTEN gene. We compared the basal VEGFA expression in LN229 with other glioma cell lines with mutant PTEN. As shown in Fig. 6c, basal VEGFA expression in LN229 is low; 4 or 9% of U251HF normalized to GAPDH or ENO1, respectively, and 2 or 6% of U87 normalized to GAPDH or ENO1, respectively. Thus VEGFA expression in LN229 apparently is under partial suppression, and can be further suppressed by PAX6 such that expression is nearly blocked, but cannot be further suppressed by increasing PTEN-blockage of PI3K/Akt signaling.
Discussion
Our data show, for the first time, that PAX6 acts as a tumor suppressor in glioma cells by suppressing angiogenesis, and that VEGFA is a target of PAX6-mediated pathways in a normoxic condition. We found that PAX6 suppression of VEGFA is under a mechanism different from the canonical mechanism involving HIF1A and VHL. Consequently, an additive suppressive effect on VEGFA expression was seen by introduction of both PAX6 and PTEN or PAX6 and PI3K inhibitor. Yet our data from this study did not identify a mediator of the PAX6 reduction of VEGFA promoter activity and consequent mRNA expression. Through transfection of different forms of PAX6 mutants, we determined that PAX6 needs its DNA-binding domain to suppress VEGFA expression, but this study did not solve the question of whether PAX6 functions directly to bind the VEGFA promoter or indirectly via another transcription factor. Data from this study reveal that PAX6 mediated a novel mechanism for regulating VEGFA expression that is independent of the PI3K/Akt-HIF1A-VHL pathway.
In this study, we used the s.c. xenograft glioma model to study the direct effect of PAX6 on angiogenesis. In this model, we can avoid complications delineating the mechanisms affecting angiogenesis in the brain, where angiogenesis can also be affected by microglia/microphage infiltration [22, 24]. Since PAX6 was found to suppress expression of the gene encoding chemokine (C–C motif) ligand 2 (CCL2) in glioma cell lines [25], using a s.c. xenograft model allowed us to exclude the microglia effect on angiogenesis in an intracranial xenograft model. With regards to the potential regulation of inflammation-induced angiogenesis by PAX6 expression in glioma cells, the intracranial xenograft model will be used. Identification of PAX6 regulation of VEGFA expression and the underlying mechanisms will provides new clues to dissect the complexity of mechanisms underlying glioma angiogenesis.
Abbreviations
- ELISA:
-
Enzyme immunometric assay
- AqRT-PCR:
-
Absolute quantitative reverse transcriptase-polymerase chain reaction
- s.c.:
-
Subcutaneous
- MI:
-
Modulated imaging
References
Simpson TI, Price DJ (2002) Pax6; a pleiotropic player in development. Bioessays 24:1041–1051
Mansouri A, Hallonet M, Gruss P (1996) Pax genes and their roles in cell differentiation and development. Curr Opin Cell Biol 8:851–857
Sander M, Neubuser A, Kalamaras J, Ee HC, Martin GR, German MS (1997) Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev 11:1662–1673
Osumi N, Shinohara H, Numayama-Tsuruta K, Maekawa M (2008) Concise review: Pax6 transcription factor contributes to both embryonic and adult neurogenesis as a multifunctional regulator. Stem Cells 26:1663–1672
Zhou YH, Tan F, Hess KR, Yung WK (2003) The expression of PAX6, PTEN, vascular endothelial growth factor, and epidermal growth factor receptor in gliomas: relationship to tumor grade and survival. Clin Cancer Res 9:3369–3375
Zhou YH, Wu X, Tan F, Shi YX, Glass T, Liu TJ, Wathen K, Hess KR, Gumin J, Lang F, Yung WK (2005) PAX6 suppresses growth of human glioblastoma cells. J Neurooncol 71:223–229
Mayes DA, Hu Y, Teng Y, Siegel E, Wu X, Panda K, Tan F, Yung WK, Zhou YH (2006) PAX6 suppresses the invasiveness of glioblastoma cells and the expression of the matrix metalloproteinase-2 gene. Cancer Res 66:9809–9817
Chang JY, Hu Y, Siegel E, Stanley L, Zhou YH (2007) PAX6 increases glioma cell susceptibility to detachment and oxidative stress. J Neurooncol 84:9–19
CBTRUS, Central Brain Tumor Registry of the United States (2005) Statistical report: primary brain tumors in the United States, 1997-2002. CBTRUS, Chicago
Norden AD, Drappatz J, Wen PY (2008) Antiangiogenic therapy in malignant gliomas. Curr Opin Oncol 20:652–661
Wiesener MS, Munchenhagen PM, Berger I, Morgan NV, Roigas J, Schwiertz A, Jurgensen JS, Gruber G, Maxwell PH, Loning SA, Frei U, Maher ER, Grone HJ, Eckardt KU (2001) Constitutive activation of hypoxia-inducible genes related to overexpression of hypoxia-inducible factor-1alpha in clear cell renal carcinomas. Cancer Res 61:5215–5222
Damert A, Machein M, Breier G, Fujita MQ, Hanahan D, Risau W, Plate KH (1997) Up-regulation of vascular endothelial growth factor expression in a rat glioma is conferred by two distinct hypoxia-driven mechanisms. Cancer Res 57:3860–3864
Guan M, Jin J, Su B, Liu WW, Lu Y (2002) Tissue factor expression and angiogenesis in human glioma. Clin Biochem 35:321–325
Hamada K, Kuratsu J, Saitoh Y, Takeshima H, Nishi T, Ushio Y (1996) Expression of tissue factor correlates with grade of malignancy in human glioma. Cancer 77:1877–1883
Poon RT, Lau CP, Ho JW, Yu WC, Fan ST, Wong J (2003) Tissue factor expression correlates with tumor angiogenesis and invasiveness in human hepatocellular carcinoma. Clin Cancer Res 9:5339–5345
Takano S, Tsuboi K, Tomono Y, Mitsui Y, Nose T (2000) Tissue factor, osteopontin, alphavbeta3 integrin expression in microvasculature of gliomas associated with vascular endothelial growth factor expression. Br J Cancer 82:1967–1973
Mukhopadhyay D, Knebelmann B, Cohen HT, Ananth S, Sukhatme VP (1997) The von Hippel-Lindau tumor suppressor gene product interacts with Sp1 to repress vascular endothelial growth factor promoter activity. Mol Cell Biol 17:5629–5639
Gomez-Manzano C, Fueyo J, Jiang H, Glass TL, Lee HY, Hu M, Liu JL, Jasti SL, Liu TJ, Conrad CA, Yung WK (2003) Mechanisms underlying PTEN regulation of vascular endothelial growth factor and angiogenesis. Ann Neurol 53:109–117
Zhou YH, Hess RK, Liu L, Linskey ME, Yung WKA (2005) Modeling prognosis for patients with malignant astrocytic gliomas: quantifying the expression of multiple genetic markers and clinical variables. Neuro Oncol 7:485–494
Wesseling P, van der Laak JA, de Leeuw H, Ruiter DJ, Burger PC (1994) Quantitative immunohistological analysis of the microvasculature in untreated human glioblastoma multiforme. Computer-assisted image analysis of whole-tumor sections. J Neurosurg 81:902–909
Cuccia DJ, Bevilacqua F, Durkin AJ, Tromberg BJ (2005) Modulated imaging: quantitative analysis and tomography of turbid media in the spatial-frequency domain. Opt Lett 30:1354–1356
Platten M, Kretz A, Naumann U, Aulwurm S, Egashira K, Isenmann S, Weller M (2003) Monocyte chemoattractant protein-1 increases microglial infiltration and aggressiveness of gliomas. Ann Neurol 54:388–392
Ishii N, Maier D, Merlo A, Tada M, Sawamura Y, Diserens AC, Van Meir EG (1999) Frequent co-alterations of TP53, p16/CDKN2A, p14ARF, PTEN tumor suppressor genes in human glioma cell lines. Brain Pathol 9:469–479
Goede V, Brogelli L, Ziche M, Augustin HG (1999) Induction of inflammatory angiogenesis by monocyte chemoattractant protein-1. Int J Cancer 82:765–770
Zhou YH, Glass T, Yung WKA (2001) PAX6 down regulate the expression of PDGFRA and MCP-1 genes in glioblastoma cells. Neuro Oncol 3:312
Acknowledgments
This work was supported in part by the Arkansas Cancer Research Center at the University of Arkansas for medical sciences, and University of California, Irvine, Committee on Research Award.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Zhou, YH., Hu, Y., Mayes, D. et al. PAX6 suppression of glioma angiogenesis and the expression of vascular endothelial growth factor A. J Neurooncol 96, 191–200 (2010). https://doi.org/10.1007/s11060-009-9963-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-009-9963-8