Abstract
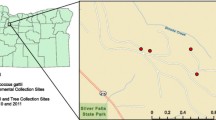
Until recently, Cryptococcus gattii was believed to be endemic in tropical and subtropical regions. To date, it has unexpectedly emerged as primary pathogen in temperate climate indicating that it has evolved and adapted to new environmental conditions including those existing in the Mediterranean area. Earlier attempts to isolate C. gattii from our environment were unsuccessful but this time, 18 years after the last environmental screening for C. neoformans, we isolated C. gattii from Eucalyptus camaldulensis in Reggio Calabria, Italy. The strains were serotype B, mating type α and were assigned to the molecular type VGI. In this study, we reported the first real environmental isolation of C. gattii in southern Italy that emphasized the observed global expansion of this yeast.


Similar content being viewed by others
References
Mc Clelland EE, Casadevall A, Eisenman HC. Pathogenesis of Cryptococcus neoformans. In: Kavanagh K, editor. New insights in medical mycology. Dordrecht: Springer; 2007. p. 131–57.
Martinez LR, Garcia-Rivera J, Casadevall A. Cryptococcus neoformans var. neoformans (serotype D) strains are more susceptible to heat than C. neoformans var. grubii (serotype A) strains. J Clin Microbiol. 2001;39:3365–7.
Kwon-Chung KJ, Boekhout T, Fell JW, Diaz M. Proposal to conserve the name Cryptococcus gattii against C. hondurianus, C. bacillisporus (Basidiomycota, Hymenomycetes, Tremellomycetidae). Taxon. 2002;51:804–6.
Springer DJ, Chaturvedi V. Projecting global occurrence of Cryptococcus gattii. Emerg Infect Dis. 2010;16:14–20.
Meyer W, Castañeda A, Jackson S, Huynh M, Castañeda E. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg Infect Dis. 2003;9:189–95.
Litvintseva AP, Thakur R, Vilgalys R, Mitchell TG. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics. 2006;172:2223–38.
Bovers M, Hagen F, Kuramae EE, Boekhout T. Six monophyletic lineages identified within Cryptococcus neoformans and Cryptococcus gattii by multi-locus sequence typing. Fungal Genet Biol. 2008;45:400–21.
Lin X, Litvintseva AP, Nielsen K, Patel S, Floyd A, Mitchell TG, Heitman J. alpha AD alpha hybrids of Cryptococcus neoformans: evidence of same-sex mating in nature and hybrid fitness. PLoS Genet. 2007;3:1975–90.
Bovers M, Hagen F, Kuramae EE, Diaz MR, Spanjaard L, Dromer F, Hoogveld HL, Boekhout T. Unique hybrids between the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii. FEMS Yeast Res. 2006;6:599–607.
Bovers M, Hagen F, Kuramae EE, Hoogveld HL, Dromer F, St-Germain G, Boekhout T. AIDS patient death caused by novel Cryptococcus neoformans x C. gattii hybrid. Emerg Infect Dis. 2008;14:1105–8.
Dixit A, Carroll SF, Qureshi ST. Cryptococcus gattii: an emerging cause of fungal disease in North America. Interdiscip Perspect Infect Dis. 2009. doi:10.1155/2009/840452.
Ellis DH, Pfeiffer TJ. Natural habitat of Cryptococcus neoformans var. gattii. J Clin Microbiol. 1990;28:1642–4.
Campisi E, Mancianti F, Pini G, Faggi E, Gargani G. Investigation in Central Italy of the possible association between Cryptococcus neoformans var. gattii and Eucalyptus camaldulensis. Eur J Epidemiol. 2003;18:357–62.
Hoang LM, Maguire JA, Doyle P, Fyfe M, Roscoe DL. Cryptococcus neoformans infections at Vancouver Hospital and Health Sciences Centre (1997–2002): epidemiology, microbiology and histopathology. J Med Microbiol. 2004;53:935–40.
Byrnes EJ, Li W, Lewit Y, Ma H, Voelz K, Ren P, Carter DA, Chaturvedi V, Bildfell RJ, May RC, Heitman J. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog. 2010. doi:10.1371/journal.ppat.1000850.
Dromer F, Mathoulin S, Dupont B. Laporte A epidemiology of cryptococcosis in France: a 9 year survey (1985–1993). French Cryptococcosis study group. Clin Infect Dis. 1996;23:82–90.
Montagna MT, Tortorano AM, Fiore L, Ingletti AM, Barbuti S. Cryptococcus neoformans var. gattii en Italie. Not I. Premier cas autochtone de méningite à sérotype B chez un sujet VIH positif. J Mycol Med. 1997;7:90–2.
Velegraki A, Kiosses VG, Pitsouni H, Toukas D, Daniilidis VD, Legakis NJ. First report of Cryptococcus neoformans var. gattii serotype B from Greece. Med Mycol. 2001;39:419–22.
Colom MF, Frasés S, Ferrer C, Jover A, Andreu M, Reus S, Sánchez M, Torres-Rodríguez JM. First case of human cryptococcosis due to Cryptococcus neoformans var. gattii in Spain. J Clin Microbiol. 2005;43:3548–50.
Solla I, Morano LE, Vasallo F, Cuenca-Estrella M. Cryptococcus gattii meningitis: observation in a Spanish patient. Enferm Infecc Microbiol Clin. 2008;26:395–6.
Montagna MT, Viviani MA, Pulito A, Aralla C, Tortorano AM, Fiore L, Barbuti S. Cryptococcus neoformans var. gtatii in Italy. Note II. Environmental investigations related to an autochthonous clinical case in Apulia. J Mycol Med. 1997;7:93–6.
Criseo G, Messina S, Chindemi G. Habitat naturali di Cryptococcus neoformans nei centri urbani di Messina e Reggio Calabria: studio sulla presenza delle varietà ‘neoformans’ e ‘gattii’. In: Proceedings of 1st Congresso Nazionale FIMUA, Firenze, Italy. 1992. p. 155.
Pernice I, Lo Passo C, Criseo G, Pernice A, Todaro-Luck F. Molecular subtyping of clinical and environmental strains of Cryptococcus neoformans variety neoformans serotype A isolated from southern Italy. Mycoses. 1998;41:117–24.
Enache-Angoulvant A, Chandenier J, Symoens F, Lacube P, Bolognini J, Douchet C, Poirot JL, Hennequin C. Molecular identification of Cryptococcus neoformans serotypes. J Clin Microbiol. 2007;45:1261–5.
Feng X, Yao Z, Ren D, Liao W. Simultaneous identification of molecular and mating types within the Cryptococcus species complex by PCR-RFLP analysis. J Med Microbiol. 2008;57:1481–90.
Kwon-Chung KJ, Polacheck I, Bennett JE. Improved diagnostic medium for separation of Cryptococcus neoformans var. neoformans (serotypes A and D) and Cryptococcus neoformans var. gattii (serotypes B and C). J Clin Microbiol. 1982;15:535–7.
Dufait R, Velho R, De Vroey C. Rapid identification of the two varieties of Cryptococcus neoformans by d-Proline. Mycoses. 1987;30:483.
Mukamurangwa P, Raes-Wuytack C, De Vroey C. Cryptococcus neoformans var. gattii can be separated from the var. neoformans by its ability o assimilate d-tryptophan. J Med Vet Mycol. 1995;33:419–20.
Müller FM, Werner KE, Kasai M, Francescani A, Chanock SJ, Walsh TJ. Rapid extraction of genomic DNA from medically important yeasts and filamentous fungi by high-speed cell disruption. J Clin Microbiol. 1998;36:1625–9.
Klein KR, Hall L, Deml SM, Rysavy JM, Wohlfiel SL, Wengenack NL. Identification of Cryptococcus gattii by use of L-canavanine glycine bromothymol blue medium and DNA sequencing. J Clin Microbiol. 2009;47:3669–72.
O’Donnell K. Fusarium and its near relatives. In: Reynolds R, Taylor JW, editors. The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. UK: CAB International; 1993. p. 225–33.
White TM, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. p. 315–21.
Kang Y, Tanaka H, Moretti ML, Mikami Y. New ITS genotype of Cryptococcus gattii isolated from an AIDS patient in Brazil. Microbiol Immunol. 2009;53:112–6.
Kwon-Chung KJ, Kwon-Chung KJ, Bennett JE, Bennett JE, Bennett JE. High prevalence of Cryptococcus neoformans var. gattii in tropical and subtropical regions. Zentralbl Bakteriol Mikrobiol Hyg [A]. 1984;257:213–8.
Sorrell TC. Cryptococcus neoformans variety gattii. Med Mycol. 2001;39:155–68.
Martins HM, Bernardo FM, Martins ML. Cryptococcus spp. associated with Eucalyptus trees in Lisboa, Portugal. In: Proceedings of 3rd meeting of the European confederation of medical mycology, Lisboa, Portugal. 1996.
Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, Diezmann S, Allen A, Stajich JE, Dietrich FS, Perfect JR, Heitman J. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437:1360–4.
Baró T, Torres-Rodríguez JM, De Mendoza MH, Morera Y, Alía C. First identification of autochthonous Cryptococcus neoformans var. gattii isolated from goats with predominantly severe pulmonary disease in Spain. J Clin Microbiol. 1998;36:458–61.
Hagen F, Van Assen S, Luijckx GJ, Boekhout T, Kampinga GA. Activated dormant Cryptococcus gattii infection in a Dutch tourist who visited Vancouver Island (Canada): a molecular epidemiological approach. Med Mycol. 2010;48:528–31.
Frasés S, Ferrer C, Sánchez M, Colom-Valiente MF. Molecular epidemiology of isolates of the Cryptococcus neoformans species complex from Spain. Rev Iberoam Micol. 2009;26:112–7.
Ngamskulrungroj P, Gilgado F, Faganello J, Litvintseva AP, Leal AL, Tsui KM, Mitchell TG, Vainstein MH, Meyer W. Genetic diversity of the Cryptococcus species complex suggests that Cryptococcus gattii deserves to have varieties. PLoS One. 2009. doi:10.1371/journal.pone.0005862.
Lindberg J, Hagen F, Laursen A, Stenderup J, Boekhout T. Cryptococcus gattii risk for tourists visiting Vancouver Island, Canada. Emerg Infect Dis. 2007;13:178–9.
Georgi A, Schneemann M, Tintelnot K, Calligaris-Maibach RC, Meyer S, Weber R, Bosshard PP. Cryptococcus gattii meningoencephalitis in an immunocompetent person 13 months after exposure. Infection. 2009;37:370–3.
Tintelnot K, Lemmer K, Losert H, Schär G, Polak A. Follow-up of epidemiological data of cryptococcosis in Austria, Germany and Switzerland with special focus on the characterization of clinical isolates. Mycoses. 2004;47:455–64.
Katsu M, Kidd S, Ando A, Moretti-Branchini ML, Mikami Y, Nishimura K, Meyer W. The internal transcribed spacer and 5.8S rRNA gene show extensive diversity among isolates of the Cryptococcus neoformans species complex. FEMS Yeast Res. 2004;4:377–88.
Ellis D, Marriott D, Hajjeh RA, Warnock D, Meyer W, Barton R. Epidemiology: surveillance of fungal infections. Med Mycol. 2000;38:173–82.
Viviani MA, Cogliati M, Esposto MC, Lemmer K, Tintelnot K, Colom Valiente MF, Swinne D, Velegraki A, Velho R. Molecular analysis of 311 Cryptococcus neoformans isolates from a 30-month ECMM survey of cryptococcosis in Europe. FEMS Yeast Res. 2006;6:614–9.
Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, Macdougall L, Boekhout T, Kwon-Chung KJ, Meyer W. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc Natl Acad Sci USA. 2004;101:17258–63.
Acknowledgments
We are very grateful to Prof. Demetrio Delfino (The Elie Metchnikoff Department, University of Messina, Italy) for generously providing us the C. neoformans H99 strain. We also thank Prof. Anna Maria Tortorano (Department of Public Health-Microbiology-Virology, School of Medicine, University of Milan, Italy) for sending us the C. gattii strains NIH444, NIH191, WM779 and WM163.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Romeo, O., Scordino, F. & Criseo, G. Environmental Isolation of Cryptococcus gattii Serotype B, VGI/MATα Strains in Southern Italy. Mycopathologia 171, 423–430 (2011). https://doi.org/10.1007/s11046-010-9389-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11046-010-9389-z