Abstract
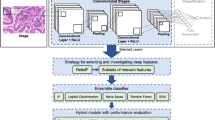
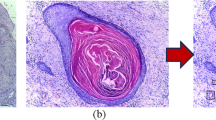
Medical image classification is an important and challenging problem, since images are usually complex, variable and the amount of data is relatively constrained. Selecting optimal sets of features and classifiers is a crucial problem in this area. In this paper it is proposed an image classification method, named Hybrid CNN Ensemble (HCNNE), based on the combination of image features extracted by convolutional neural networks (CNN) and local binary patterns (LBP). The features are subsequently used to build an ensemble of multiple classifiers. More specifically, the Euclidean distance between LBP feature vectors of each training class and the confidence of CNN features classified by support vector machines are employed to compose the input of a multilayer perceptron classifier. Finally, these features are also used as input to other classifiers to compose the final voting ensemble. This approach achieved an accuracy similar to those of other state-of-the-art methods in texture classification and showed an improvement of 10% over the previously reported identification of a group of odontogenic oral cyst histological images, at a low computational cost. Three major contributions are presented here: 1) the combination of low and high level features assigning weights based on the confidence of the features for texture recognition; 2) the combination of automatically learned deep features with LBP by a multilayer perceptron based on the feature confidences; 3) state-of-the-art results are obtained in the odontogenic cyst categorization problem.
















Similar content being viewed by others
Data Availability
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Notes
Here ‘level’ means the complexity of the feature, i.e. how much information about the image it holds.
References
Alipanahi B, Delong A, Weirauch MT, Frey BJ (2015) Predicting the sequence specificities of DNA-and RNA-binding proteins by deep learning. Nat Biotechnol 33:831–838
Anwar F, Attallah O, Ghanem N, Ismail MA (2020) Automatic breast cancer classification from histopathological images. In: 2019 International conference on advances in the emerging computing technologies (AECT), pp 1–6
Attallah O (2022) ECG-Biconet: an ECG-based pipeline for COVID-19 diagnosis using Bi-Layers of deep features integration. Comput Biol Med 142:105210
Attallah O, Sharkas M (2021) GASTRO-CADX: a three stages framework for diagnosing gastrointestinal diseases. PeerJ Comput Sci 7:e423
Attallah O, Sharkas M (2021) Intelligent dermatologist tool for classifying multiple skin cancer subtypes by incorporating manifold radiomics features categories. Contrast Media Mol Imaging
Attallah O, Anwar F, Ghanem NM, Ismail MA (2021) Histo-CADx: duo cascaded fusion stages for breast cancer diagnosis from histopathological images. PeerJ Comput Sci 7:e493
Bacanin N, Zivkovic M, Al-Turjman F, Venkatachalam K, Trojovskỳ P, Strumberger I, Bezdan T (2022) Hybridized sine cosine algorithm with convolutional neural networks dropout regularization application. Sci Rep 12:1–20
Bousetouane F, Morris B (2015) Off-the-shelf CNN features for fine-grained classification of vessels in a maritime environment. In: International symposium on visual computing, pp 379–388
Breiman L (2001) Random forests. Mach Learn 45:5–32
Bruna J, Mallat S (2013) Invariant scattering convolution networks. IEEE Trans Pattern Anal Mach Intell 35:1872–1886
Burges CJ (1998) A tutorial on support vector machines for pattern recognition. Data Mining Knowl Discov 2:121–167
Cimpoi M, Maji S, Kokkinos I, Mohamed S, Vedaldi A (2014) Describing textures in the wild. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 3606–3613
Cimpoi M, Maji S, Kokkinos I, Vedaldi A (2016) Deep filter banks for texture recognition, description, and segmentation. Int J Comput Vis 118:65–94
Cortes C, Vapnik V (1995) Support-vector networks. Mach Learn 20:273–297
Dalal N, Triggs B (2005) Histograms of oriented gradients for human detection. In: 2005 IEEE Computer society conference on computer vision and pattern recognition (CVPR’05), pp 886–893
Deng J, Dong W, Socher R, Li L -J, Li K, Fei-Fei L (2009) Imagenet: a large-scale hierarchical image database. In: 2009 IEEE Conference on computer vision and pattern recognition, pp 248–255
Fix E, Hodges JL (1989) Discriminatory analysis. Nonparametric discrimination: consistency properties. Int Stat Rev/Revue Internationale de Statistique 57:238–247
Florindo JB, Bruno OM, Landini G (2017) Morphological classification of odontogenic keratocysts using Bouligand–Minkowski fractal descriptors. Comput Biol Med 81:1–10
Forcén J, Pagola M, Barrenechea E, Bustince H (2019) Combination of features through weighted ensembles for image classification. Appl Soft Comput 84:105698
Fouad S, Randell D, Galton A, Mehanna H, Landini G (2017) Unsupervised morphological segmentation of tissue compartments in histopathological images. PloS one 12:e0188717
Goodfellow I, Bengio Y, Courville A (2016) Deep learning. MIT Press, Cambridge
Guo Z, Zhang L, Zhang D (2010) A completed modeling of local binary pattern operator for texture classification. IEEE Trans Image Process 19:1657–1663
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Ho Y -C, Pepyne DL (2002) Simple explanation of the no-free-lunch theorem and its implications. J Optim Theory Appl 115:549–570
Kannala J, Rahtu E (2012) Bsif: binarized statistical image features. In: Proceedings of the 21st international conference on pattern recognition (ICPR2012), pp 1363–1366
Kumar A, Kim J, Lyndon D, Fulham M, Feng D (2016) An ensemble of fine-tuned convolutional neural networks for medical image classification. IEEE J Biomed Health Inform 21:31–40
Landini G (2006) Quantitative analysis of the epithelial lining architecture in radicular cysts and odontogenic keratocysts. Head Face Med 2:4
Landini G, Othman IE (2004) Architectural analysis of oral cancer, dysplastic, and normal epithelia. Cytometry Part A: J Int Soc Anal Cytol 61:45–55
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444
Lin H -T, Lin C -J, Weng RC (2007) A note on Platt’s probabilistic outputs for support vector machines. Mach Learn 68:267–276
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, Van Der Laak JA, Van Ginneken B, Sánchez C I (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88
Liu L, Fieguth P, Guo Y, Wang X, Pietikäinen M (2017) Local binary features for texture classification: taxonomy and experimental study. Pattern Recogn 62:135–160
Malakar S, Ghosh M, Bhowmik S, Sarkar R, Nasipuri M (2020) A GA based hierarchical feature selection approach for handwritten word recognition. Neural Comput Appl 32:2533–2552
Ojala T, Pietikainen M, Maenpaa T (2002) Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans Pattern Anal Mach Intell 24:971–987
Platt J, et al. (1999) Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. Advances in Large Margin Classifiers 10:61–74
Quan Y, Xu Y, Sun Y, Luo Y (2014) Lacunarity analysis on image patterns for texture classification. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 160–167
Ragab DA, Attallah O (2020) FUSI-CAD: coronavirus (COVID-19) diagnosis based on the fusion of CNNs and handcrafted features. PeerJ Comput Sci 6:e306
Ragab DA, Sharkas M, Attallah O (2019) Breast cancer diagnosis using an efficient CAD system based on multiple classifiers. Diagnostics 9:165
Rao CR (1948) The utilization of multiple measurements in problems of biological classification. J R Stat Soc Ser B (Methodol) 10:159–203
Ren Y, Zhang L, Suganthan PN (2016) Ensemble classification and regression-recent developments, applications and future directions. IEEE Comput Intell Mag 11:41–53
Rokach L (2010) Ensemble-based classifiers. Artif Intell Rev 33:1–39
Timofte R, Van Gool LA (2012) Training-free classification framework for textures, writers, and materials. In: BMVC, p 14
UIUC Dataset http://www-cvr.ai.uiuc.edu/poncegrp/data/index.html. Last accessed 06-2021
UMD Dataset http://cfar.umd.edu/_fer/High-resolution-database/hrdatabase.htm. Last accessed 06-2021
Wei J, He J, Chen K, Zhou Y, Tang Z (2017) Collaborative filtering and deep learning based recommendation system for cold start items. Expert Syst Appl 69:29–39
Wolpert DH (1996) The lack of a priori distinctions between learning algorithms. Neural Comput 8:1341–1390
Xu Y, Yang X, Ling H, Ji H (2010) A new texture descriptor using multifractal analysis in multi-orientation wavelet pyramid. In: 2010 IEEE Computer society conference on computer vision and pattern recognition, pp 161–168
Xu Y, Huang S, Ji H, Fermüller C (2012) Scale-space texture description on sift-like textons. Comput Vis Image Underst 116:999–1013
Acknowledgements
Marina Rocha gratefully acknowledges the support of the National Council for Scientific and Technological Development, CNPq (Grant #121791/2019-0). Joao Florindo gratefully acknowledges the financial support of São Paulo Research Foundation (FAPESP) (Grant #2020/01984-8) and from National Council for Scientific and Technological Development, Brazil (CNPq) (Grants #306030/2019-5 and #423292/2018-8).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors certify that they have no known affiliations with or involvement in any organization or entity with any interest in the subject matter discussed in this paper.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rocha, M.M.M., Landini, G. & Florindo, J.B. Medical image classification using a combination of features from convolutional neural networks. Multimed Tools Appl 82, 19299–19322 (2023). https://doi.org/10.1007/s11042-022-14206-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-022-14206-y