Abstract
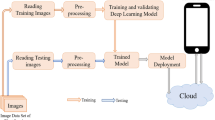
The deep learning techniques have been playing an important role in the identification and classification problems such as diseases in medical science, marketing in the industry, manufacturing in engineering, and identification in plant taxonomy science. Fruit identification and its family classification is among one of the areas that needs more emphasis for the sake of automation. With this inspiration, fruit images for 52 species belonging to four different families (Apiaceae, Brassicaceae, Asteraceae, and Apocynaceae) have been used in this study to build a deep learning analysis dataset. Further, the dataset has been augmented to 3800 images, divided to 2660 images for training and 1440 for testing, and different 14 fruit images belonging to the same families have been used for prediction of the testing module. A novel Convolution Neural Network (CNN) model architecture has been proposed to extract the fruit features, classify each image with its family, and use the trained model to predict that the new fruits belong to the same four families. The maximum accuracy obtained for the training and testing module was 99.82%. The prediction for this module succeeded by 93% since all fruits’ success predicted was attained except one from the family number 2 (Brassicaceae). The same dataset was applied to two different models to evaluate our proposed model, the Deep learning model, aka Residual Neural Network, 20 layers (ResNet-20), and Support Vector Machine (SVM). The proposed CNN model achieved higher accuracy and efficiency than the ResNet-20 and SVM.









Similar content being viewed by others
References
Ahmad M, Qadir MA, Rahman A, Zagrouba R, Alhaidari F et al (2020) Enhanced query processing over semantic cache for cloud based relational databases. J Ambient Intell Humaniz Comput. https://doi.org/10.1007/s12652-020-01943-x
Alhaidari F, Rahman A, Zagrouba R (2020) Cloud of things: architecture, applications and challenges. J Ambient Intell Humaniz Comput. https://doi.org/10.1007/s12652-020-02448-3
Alotaibi SM, Rahman A, Basheer MI, Khan MA (2021) Ensemble machine learning based identification of pediatric epilepsy. Comput Mater Continua 68(1):149–165
Biswas S, Dash S, Acharya S (2018) Firefly algorithm based multilingual named entity recognition for Indian languages. In: Proc Luhach A, Singh D, Hsiung PA, Hawari K, Lingras P, Singh P (eds) Advanced Informatics for Computing Research. ICAICR Communications in Computer and Information Science, vol 955. Springer, Singapore, pp 540–552
Dash S, Behera R (2016) Sampling based hybrid algorithms for imbalanced data classification. Int J Hybrid Intell Syst 13(1):77–86. https://doi.org/10.3233/HIS-160226
Dash S, Patra BN (2016) Genetic diagnosis of cancer by evolutionary fuzzy-based neural network ensemble. Int J Appl Res Bioinf 6(1):1–20
Dash S, Thulasiram R, Thulasiram P (2019) Modified firefly based meta-search algorithm for feature selection: a predictive model for medical data. Int J Swarm Intell 10(2):1–20
Dash S, Abraham A, Luhach AK, Mizera-Pietraszko J, Rodrigues JJPC (2019) Hybrid chaotic firefly decision-making model for Parkinson’s disease diagnosis. Int J Distrib Sens Netw 16(12):1–18
Dash S, Biswas S, Banerjee D, Rahman A (2019) Edge and fog computing in healthcare – a review. Scalable Comput 20(2):191–206
Dileep MR (2019) AyurLeaf: a deep learning approach for classification of medicinal plants. In: Proc TENCON 2019–2019 IEEE Region 10 Conference (TENCON), pp 319–323
Grillo O, Blangiforti S, Venora G (2017) Wheat landraces identification through glumes image analysis. Comput Electron Agric 141:223–231
Gyires-Tóth BP, Osváth M, Papp D, Szucs G (2019) Deep learning for plant classification and content-based image retrieval. Cybern Inf Technol 19(1):88–100
Haupt J, Kahl S, Kowerko D, Eibl M (2018)Large-scale plant classification using deep convolutional neural networks. In: Proc CEUR Workshop, vol 2125, pp 1–7
He K, Thang X, Ren S, Sun J (2015) Deep residual learning for image recognition. In: Proc IEEE conference on computer vision and pattern recognition, pp 770–778
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proc IEEE Computer Society Conference on Computer Vision and Pattern Recognition, pp 770–778. https://doi.org/10.1109/CVPR.2016.90
Hossain MS, Al-Hammadi M, Muhammad G (2019) Automatic fruit classification using deep learning for industrial applications. In: IEEE Trans Ind Inform 15(2):1027–1034. https://doi.org/10.1109/TII.2018.2875149
Huang G, Liu Z, Maaten LV, Weinberger KQ (2016) Densely connected convolutional Networks. In: Proc IEEE conference on computer vision and pattern recognition, vol 1, no 2, p 3
Jana BK, Mukherjee SK (2012) Diversity of cypselar features of seven species of the genus crepis L. in compositae. Indian J Fundam Appl Life Sci 2(1):51–58
Johannes A, Picon A, Alvarez-Gila A, Echazarra J, Rodriguez-Vaamonde S et al (2017) Automatic plant disease diagnosis using mobile capture devices, applied on a wheat use case. Comput Electron Agric 138:200–209
Justine B, Samuel F, Jérôme T, Pierre-Luc S (2019) Convolutional neural networks for the automatic identification of plant diseases. Front Plant Sci 10:941–956
Kaya A, Keceli AS, Catal C, Yalic HY, Temucin H et al (2019) Analysis of transfer learning for deep neural network-based plant classification models. Comput Electron Agric 158(1):20–29
Khan MA, Abbas S, Atta A, Ditta A, Alquhayz H et al (2020) Intelligent cloud based heart disease prediction Ssystem empowered with supervised machine learning. Comput Mater Continua 65(1):139–151
Kingma DP, Ba JL (2015) ADAM: a method for stochastic optimization. In: Proc International Conference on Learning Representations (ICRL 2015), pp 1–15
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: Proc. 25th International Conference on Neural Information Processing Systems (NIPS’12), vol 1. Curran Associates Inc. USA, pp 1097–1105
Le TL, Dng DN, Vu H, Nguyen TN (2015) Mica at LifeCLEF 2015: multi-organ plant Identification. In: Proc Working Notes of CLEF 2015 Conference
Lee JW, Yoon YC (2019)Fine-grained plant identification using wide and deep learning model 1. In: Proc International Conference on Platform Technology and Service (PlatCon 2019), pp 1–5. https://doi.org/10.1109/PlatCon.2019.8669407
Mahmud M, Rahman A, Lee M, Choi J (2020)Evolutionary-based image encryption using RNA codons truth table. Opt Laser Technol 121(1):1–8
Naseem MT, Qureshi IM, Rahman A, Muzaffar MZ (2020) Robust and fragile watermarking for medical images using redundant residue number system and chaos. Neural Netw World 30(3):177–192
Panda M, Dash S (2019) A Framework for testing object oriented programs using hybrid nature inspired algorithms. In: Proc A. K. Luhach et al (Eds.): ICAICR 2018, CCIS 955. Springer Nature, Singapore, pp 1–9. https://doi.org/10.1007/978-981-13-3140-4
Patra BN, Dash S (2016) A FRGSNN hybrid feature selection combining FRGS filter and GSNN wrapper. Int J Latest Trends Eng Technol 7(2):8–15
Picon A, Alvarez-Gila A, Seitz M, Ortiz-Barredo A, Echazarra J et al (2019) Deep convolutional neural networks for mobile capture device-based crop disease classification in the wild. Comput Electron Agric 161:280–290
Rahman A (2019) Optimum information embedding in digital watermarking. J Intell Fuzzy Syst 37(1):553–564
Rahman A (2019) Memetic computing based numerical solution to Troesch problem. J Intell Fuzzy Syst 37(1):1545–1554
Rahman A (2020)GRBF-NN based ambient aware realtime adaptive communication in DVB-S2. J Ambient Intell Humaniz Comput. https://doi.org/10.1007/s12652-020-02174-w
Rahman A, Dash S (2017) Big data analysis for teacher recommendation using data mining techniques. Int J Control Theory Appl 10(18):95–105
Rahman A, Sultan K, Aldhafferi N, Alqahtani A, Mahmud M (2018) Reversible and fragile watermarking for medical images. Comput Math Methods Med Article ID 3461382, 7 pages. https://doi.org/10.1155/2018/3461382
Rahman A, Sultan K, Musleh D, Aldhafferi N, Alqahtani A, Mahmud M (2018) Robust and fragile medical image watermarking: a joint venture of coding and chaos theories. J Healthc Eng Article ID 8137436, 11 pages. https://doi.org/10.1155/2018/8137436
Rahman A, Dash S, Luhach AK, Chilamkurti N, Baek S et al (2019) A neuro-fuzzy approach for user behavior classification and prediction. J Cloud Comput 8:17
Rahman A, Dash S, Luhach AK (2020) Dynamic MODCOD and power allocation in DVB-S2: a hybrid intelligent approach. Telecommun Syst. https://doi.org/10.1007/s11235-020-00700-x
Rahman A, Sultan K, Naseer I, Majeed R, Musleh D et al (2021) Supervised machine learning-based prediction of COVID-19. Comput Mater Continua 69(1):21–34
Rehman A, Athar A, Khan MA, Abbas S, Rahman A et al (2020) Modelling, simulation, and optimization of diabetes type II prediction using deep extreme learning machine. J Ambient Intell Smart Environ 12(2):125–138
Reyes AK, Caicedo JC, Camargo JE (2015)Fine-tuning deep convolutional networks for plant cecognition. In: Proc Working Notes of CLEF
Sabzi S, Abbaspour-Gilandeh Y, García-Mateos G (2018) A new approach for visual identification of orange varieties using neural networks and metaheuristic algorithms. Inform Process Agric 5(1):162–172. https://doi.org/10.1016/j.inpa.2017.09.002
Shi Y, Wei Z, Ling H, Wang Z, Shen J, Li P. Person retrieval in surveillance videos via deep attribute mining and reasoning. In: IEEE Trans Multimed. https://doi.org/10.1109/TMM.2020.3042068
Singh R, Dash S, Biswas S, Deka B (2020) Mobile technology solution for COVID-19. In: Proc. Fadi Al-Turjaman et al. (eds) Emerging Technologies for battling COVID-19 applications and innovations. Springer, Berlin, ISBN: 978-030-60038-9
Sungbin C (2015) Plant identification with deep convolutional neural network: SNUMedinfo at LifeCLEF plant identification task 2015. In: Proc Working Notes of CLEF
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2015) Rethinking the incpetion architecture of computer vision. In: Proc IEEE Conference on Computer Vision and Pattern Recognition, pp 2818–2826
Tan JW, Chang SW, Binti KA, Yap HJ, Yong KT (2018) Deep learning for plant species classification using leaf vein morphometric. IEEE/ACM Trans Comput Biol Bioinform 5963(c). https://doi.org/10.1109/TCBB.2018.2848653
Dash S, Behera R (2016) Sampling based hybrid algorithms for imbalanced data classification. Int J Hybrid Intell Syst 13(1):77–86. https://doi.org/10.3233/HIS-160226
Xiong Z, Yuan Y, Wang Q (2021) Adaptively Selecting Key Local Features for RGB-D Scene Recognition,“. IEEE Trans Image Process 30:2722–2733. https://doi.org/10.1109/TIP.2021.3053459
Zagrouba R, Khan MA, Rahman A, Saleem MA, Mushtaq MF et al (2021) Modelling and simulation of COVID-19 outbreak prediction using supervised machine learning. Comput Mater Continua 66(6):2397–2407
Zaman G, Mahdin H, Hussain K, Rahman A, Abawajy J, Mostafa SA (2021) "An Ontological Framework for Information Extraction From Diverse Scientific Sources,“. IEEE Access 9:42111–42124
Zhang H, He G, Peng J, Kuang Z, Fan J (2018) Deep learning of path-based tree classifiers for large-scale plant species identification. In: Proc IEEE 1st Conference on Multimedia Information Processing and Retrieval, MIPR, pp 25–30. https://doi.org/10.1109/MIPR.2018.00013
Zhang C, Wang Q, Li X (2021) Towards a unified framework for license plate detection, tracking, and recognition in real-world traffic videos. Neurocomputing 449:189–206
Zheng Y, Zhu Q, Huang M, Guo Y, Qin J (2017) Maize and weed classification using color indices with support vector data description in outdoor fields. Comput Electron Agric 141:215–222
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ibrahim, N.M., Gabr, D.G.I., Rahman, Au. et al. A deep learning approach to intelligent fruit identification and family classification. Multimed Tools Appl 81, 27783–27798 (2022). https://doi.org/10.1007/s11042-022-12942-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-022-12942-9