Abstract
Background
Quantitative real-time PCR (qPCR) is a highly reliable method for validating gene expression data in molecular studies due to its sensitivity, specificity, and efficiency. To ensure accurate qPCR results, it’s essential to normalize the expression data using stable reference genes.
Methods
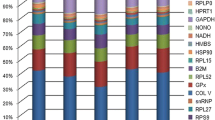
This study aimed to identify suitable reference genes for qPCR studies in goats by evaluating 18 candidate reference genes (ACTB, BACH1, B2M, GAPDH, HMBS, HPRT1, PGK1, PPIA, PPIB, RPLP0, RPL19, RPS9, RPS15, RPS28, SDHA, TBP, UXT, and YWHAZ) in 10 different caprine tissues (heart, intestine, kidney, liver, lung, muscle, rumen, skin, spleen, and testis). An integrated tool called RefFinder, which incorporates various algorithms like NormFinder, GeNorm, BestKeeper, and ΔCt, was used to assess the stability of expression among these genes.
Results
After thorough analysis, ACTB, PPIB, and B2M emerged as the most stable reference genes, while RPL19, RPS15, and RPS9 were found to be the least stable. The suitability of the selected internal control genes was further validated through target gene analysis, confirming their efficacy in ensuring accurate gene expression profiling in goats.
Conclusion
The study determined that the geometric average of ACTB, PPIB, and B2M creates an appropriate normalization factor for gene expression studies in goat tissues.

Similar content being viewed by others
Data availability
No datasets were generated or analysed during the current study.
References
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3(6):1101–1108
Sonowal J, Patel CL, Dev K, Singh R, Barkathullah N, Malla WA, Mishra B (2022) Selection and validation of suitable reference gene for qPCR gene expression analysis in lamb testis cells under sheep pox virus infection. Gene 831:146561
Kaur R, Ahlawat S, Choudhary V, Kumari A, Kumar A, Kaur M, Vijh RK (2023) Validation of stable reference genes in peripheral blood mononuclear cells for expression studies involving vector-borne haemoparasitic diseases in bovines. Ticks Tick Borne Dis 14(4):102168
Herath S, Dai H, Erlich J, Au AY, Taylor K, Succar L, Endre ZH (2020) Selection and validation of reference genes for normalisation of gene expression in ischaemic and toxicological studies in kidney disease. PLoS ONE 15(5):e0233109
Stephen AB, Vladimir B, Jeremy AG, Jan H, Jim H, Mikael K, Reinhold M, Tania N, Michael WF, Gregory LS, Jo V, Carl TW (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55(4):611–622
Nygard AB, Jørgensen CB, Cirera S, Fredholm M (2007) Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR. BMC Mol Biol 8(1):1–6
Zhang Y, Zhang XD, Liu X, Li YS, Ding JP, Zhang XR, Zhang YH (2013) Reference gene screening for analyzing gene expression across goat tissue. Asian-Australas J Anim Sci 26(12):1665–1671
Vasu M, Ahlawat S, Choudhary V, Kaur R, Arora R, Sharma R, Singh MK (2023) Identification and validation of stable reference genes for expression profiling of target genes in diverse ovine tissues. Gene 148067.
Xie F, Wang J, Zhang B (2023) RefFinder: a web-based tool for comprehensively analyzing and identifying reference genes. Funct Integr Genomics 23(2):1–5
Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64(15):5245–5250
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7):1–12
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett 26:509–515
Silver N, Best S, Jiang J, Thein SL (2006) Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol 7(1):1–9
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3-new capabilities and interfaces. Nucleic Acids Res 40 (15), e115
Coelho TC, Chalfun-Junior A, Barreto HG, Duarte MDS, Garcia BDO, Teixeira PD, Ladeira MM (2022) Reference gene selection for quantitative PCR in liver, skeletal muscle, and jejunum of Bos indicus cattle. Rev Bras De Zootec 51
Gothel SF, Marahiel MA (1999) Peptidyl-prolyl cis-trans isomerases, a superfamily of ubiquitous folding catalysts. Cell Mol Life Sci 55:423–436
Dydensborg AB, Herring E, Auclair J, Tremblay E, Beaulieu JF (2006) Normalizing genes for quantitative RT-PCR in differentiating human intestinal epithelial cells and adenocarcinomas of the colon. Am J Physiol Gastrointest 290(5):G1067–G1074
Wu X, Yang W, Ren Z, Cheng J, Luo Y, Wang Y, Yang Z, Yao X, Zhao W, Li Y, Tang K (2021) Reference gene screening for analyzing gene expression in the heart, liver, spleen, lung and kidney of forest musk deer. J Vet Med Sci 83(11):1750–1759
Zhao J, Wang C, Zhang L, Lei A, Wang L, Niu L, Zhong T (2021) Genome-wide identification of reference genes for reverse transcription quantitative PCR in goat rumen. Animals 11(11):3137
Zhao L, Yang H, LiX, Zhou Y, Liu T, Zhao Y (2022) Transcriptome-based selection and validation of optimal reference genes in perirenal adipose developing of goat (Capra hircus). Front Vet Sci 9:1055866
Zhang J, Deng C, Li J, Zhao Y (2020) Transcriptome-based selection and validation of optimal house-keeping genes for skin research in goats (Capra hircus). BMC Genom 21:1–16
Zhu W, Lin Y, Liao H, Wang Y (2015) Selection of reference genes for gene expression studies related to intramuscular fat deposition in Capra hircus skeletal muscle. PLoS ONE 10(3):e0121280
Xu Q, Lin S, Zhu J, Wang Y, Lin Y (2018) The expression stability analysis of reference genes in the process of goat intramuscular preadipocytes differentiation in goat. Acta Vet et Zootech Sin 49(5):907–918
Zhou Y, Li X, Zhang X, Li M, Luo N, Zhao Y (2023) Screening of candidate housekeeping genes in uterus caruncle by RNA-Sequence and qPCR analyses in different stages of goat (Capra hircus). Animals 13(12):1897
Luo N, Zhou Y, Chen X, Zhao Y, HuY (2024) Screening the optimal housekeeping genes (HKGs) of placenta tissues by RNA-sequence and qRT-PCR throughout gestation in goat (Capra hircus). Gene 895:147966
Sahu AR, Wani SA, Saxena S, Rajak KK, Chaudhary D, Sahoo AP, Khanduri A, Pandey A, Mondal P, Malla WA, Khan RIN (2018) Selection and validation of suitable reference genes for qPCR gene expression analysis in goats and sheep under Peste Des petits ruminants virus (PPRV), lineage IV infection. Sci Rep 8(1):1–11
Aziziyan A, Sadeghi M, Ganjkhanlou M, Bahnamiri HZ (2020) Reference gene selection in adipose and muscle tissues of fat-tailed Lori-Bakhtiari lambs. Iran J Vet Med 14(3)
Lozano-Villegas KJ, Rodriguez-HernandezR, Herrera-Sanchez MP, Uribe-Garcia HF, Naranjo-Gomez JS, Otero-Arroyo RJ, Rondon-Barragan IS (2021) Identification of reference genes for expression studies in the whole-blood from three cattle breeds under two states of livestock weather safety. Animals 11(11):3073
Caetano LC, Miranda-Furtado CL, Batista LA, Pitangui-Molina CP, Higa TT, Padovan CC, de Sá Rosa-e-Silva ACJ (2019) Validation of reference genes for gene expression studies in bovine oocytes and cumulus cells derived from in vitro maturation. Anim Reprod 16(2):290–296
Funding
This work was financially supported by the ICAR-Consortium Research Platform- Genomics.
Author information
Authors and Affiliations
Contributions
S.A.: conceptualization, generation of the manuscript. M.V.: data curation, formal analyses. V.C., M.M., and M.K.S.: resources. R.A. and R.S.: formal analyses. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Ethical approval
This work was approved by the Institutional Animal Ethics Committee (IAEC) of ICAR-National Bureau of Animal Genetics Resources (F. No. NBAGR/IAEC/2017, dated 21.01.2017).
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ahlawat, S., Vasu, M., Choudhary, V. et al. Comprehensive evaluation and validation of optimal reference genes for normalization of qPCR data in different caprine tissues. Mol Biol Rep 51, 268 (2024). https://doi.org/10.1007/s11033-024-09268-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-024-09268-0