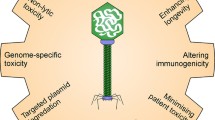
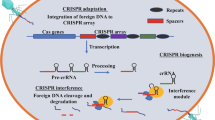
Abstract
Bacteriophages (phages) are viruses that mainly infect bacteria and are ubiquitously distributed in nature, especially to their host. Phage engineering involves nucleic acids manipulation of phage genome for antimicrobial activity directed against pathogens through the applications of molecular biology techniques such as synthetic biology methods, homologous recombination, CRISPY-BRED and CRISPY-BRIP recombineering, rebooting phage-based engineering, and targeted nucleases including CRISPR/Cas9, zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs). Management of bacteria is widely achieved using antibiotics whose mechanism of action has been shown to target both the genetic dogma and the metabolism of pathogens. However, the overuse of antibiotics has caused the emergence of multidrug-resistant (MDR) bacteria which account for nearly 5 million deaths as of 2019 thereby posing threats to the public health sector, particularly by 2050. Lytic phages have drawn attention as a strong alternative to antibiotics owing to the promising efficacy and safety of phage therapy in various models in vivo and human studies. Therefore, harnessing phage genome engineering methods, particularly CRISPR/Cas9 to overcome the limitations such as phage narrow host range, phage resistance or any potential eukaryotic immune response for phage-based enzymes/proteins therapy may designate phage therapy as a strong alternative to antibiotics for combatting bacterial antimicrobial resistance (AMR). Here, the current trends and progress in phage genome engineering techniques and phage therapy are reviewed.







Similar content being viewed by others
Data Availability
Not applicable.
Abbreviations
- AIDS:
-
Acquired immunodeficiency syndrome
- AMR:
-
Antimicrobial resistance
- CRISPY-BRED:
-
CRISPR/Cas9-bacteriophage recombineering with electroporated DNA
- CRISPY-BRIP:
-
CRISPR/Cas9-bacteriophage recombineering with infectious particles
- CRISPR-plasmid:
-
Clustered regularly interspaced short palindromic repeats-plasmid
- CRISPR-Cas:
-
Clustered regularly interspaced short palindromic repeats-CRISPR associated proteins
- CRISPR/Cas9:
-
Clustered regularly interspaced short palindromic repeats/Cas9
- DNA:
-
Deoxyribonucleic acid
- DSBs:
-
DNA double-strand breaks
- dsDNA:
-
Double-stranded DNA
- EPA:
-
Environmental protection agency
- FDA:
-
Food and drug administration
- FQs:
-
Fluoroquinolones
- HDR:
-
Homology-directed repair
- HIV:
-
Human immunodeficiency virus
- HR:
-
Homologous recombination
- MDR Bacteria:
-
Multidrug-resistant bacteria
- NHEJ:
-
Nonhomologous end joining
- ORFs:
-
Open reading frames
- RNA:
-
Ribonucleic acid
- ssDNA:
-
Single-stranded DNA
- TALENs:
-
Transcription activator-like effector nucleases
- tRNA:
-
Transfer RNA
- UN:
-
United Nations
- USA:
-
United States of America
- ZFNs:
-
Zinc-finger nucleases
References
Abdelsattar AS, Dawoud A, Makky S, Nofal R, Aziz RK, El-Shibiny A (2021) Bacteriophages: from isolation to application. Curr Pharm Biotechnol 22. https://doi.org/10.2174/1389201022666210426092002
Coffey A (2016) Bacteriophage and their lysins for elimination of infectious bacteria: review article bacteriophage and their lysins for elimination of infectious bacteria. no May 2009. https://doi.org/10.1111/j.1574-6976.2009.00176.x
Scattolini S et al (2021) Characterization and in vitro efficacy against listeria monocytogenes of a newly isolated bacteriophage, ɸizsam-1. Microorganisms 9(4). https://doi.org/10.3390/microorganisms9040731
Tao P (2019) Genetic Engineering of Bacteriophages against Infectious Diseases. no May 10:1–12. https://doi.org/10.3389/fmicb.2019.00954
Mutalik VK, Arkin AP (2022) A phage Foundry Framework to systematically develop viral countermeasures to combat antibiotic-resistant bacterial pathogens. iScience 25(4):104121. https://doi.org/10.1016/j.isci.2022.104121
Shaidullina A, Harms A (2023) Toothpicks, logic, and next-generation sequencing_ systematic investigation of bacteriophage-host interactions. Curr Opin Microbiol 70:102225. https://doi.org/10.1016/j.mib.2022.102225
Hassan AY, Lin JT, Ricker N, Anany H (2021) The age of phage: friend or foe in the new dawn of therapeutic and biocontrol applications? Pharmaceuticals 14(3):1–36. https://doi.org/10.3390/ph14030199
Ryu S (2021) “Grand Challenges in Phage Biology,” Front. Microbiol, vol. 12, no. July, pp. 2019–2022, https://doi.org/10.3389/fmicb.2021.715039
Azam AH (2021) “Bacteriophage Technology and Modern Medicine,”
Ganesh SK, Subathra Devi C (2023) Molecular and therapeutic insights of rapamycin: a multi-faceted drug from Streptomyces hygroscopicus. Mol Biol Rep 50(4):3815–3833. https://doi.org/10.1007/s11033-023-08283-x
Alebouyeh M et al (2023) Intestinal colonization of vancomycin-resistant Enterococcus in children admitted to Mofid children’s hospital intensive care unit at admission and at discharge. Mol Biol Rep 3271–3281. https://doi.org/10.1007/s11033-022-08196-1
Jain H, Chahal S, Singh I, Sain SK, Siwach P (2023) The rising threat of geminiviruses: molecular insights into the disease mechanism and mitigation strategies. Mol Biol Rep 50(4):3835–3848. https://doi.org/10.1007/s11033-023-08266-y
Cui Z, Xu Z, Wei Y, Zhang Q, Qin K, Ji X (2021) Characterization and genome analysis of a Novel Mu-like phage VW-6B isolated from the Napahai Plateau Wetland of China. Curr Microbiol 78(1):150–158. https://doi.org/10.1007/s00284-020-02277-9
Tesson F, Bernheim A (2023) Synergy and regulation of antiphage systems: toward the existence of a bacterial immune system ? Curr Opin Microbiol 71:102238. https://doi.org/10.1016/j.mib.2022.102238
John AM, Mercer DK (2023) Antimicrobial resistance: a biochemical society position statement. no February:33–38
Dewanggana MN, Evangeline C, Ketty MD, Waturangi DE, Yogiara, Magdalena S (2022) Isolation, characterization, molecular analysis and application of bacteriophage DW-EC to control enterotoxigenic Escherichia coli on various foods. Sci Rep 12(1):1–10. https://doi.org/10.1038/s41598-021-04534-8
Strathdee SA, Hatfull GF, Mutalik VK, Schooley RT (2023) Ll phage therapy: from biological mechanisms to future directions. Cell 186(1):17–31. https://doi.org/10.1016/j.cell.2022.11.017
Roszak M, Jabłon J (2022) “Bacteriophage – Ciprofloxacin Combination Effectiveness,” vol. 28, no. 6, pp. 613–622, https://doi.org/10.1089/mdr.2021.0324
Lopez-luis BA, Ponce-de-leo A, Ortiz-brizuela E, Leal-vega FJ, Tovar-caldero YE (2022) Bobadilla-del-valle, “Risk factors Associated with failure of Linezolid Therapy in Vancomycin-Resistant Enterococcus faecium bacteremia. 28(6):744–749. https://doi.org/10.1089/mdr.2021.0333
Chen J et al (2022) Different Effects of Antibiotics on Klebsiella pneumoniae and Escherichia coli Resistance Induced by Antibiotics. 28(6):660–669. https://doi.org/10.1089/mdr.2021.0326
Ahmed BT, Mokhtar B, Yahia B, Wassila C (2023) “Antibiotic resistance: A Global Public Health Crisis and current strategies for Antibiotic Resistance : A Global Public Health Crisis and current strategies for combatting it,” no. April,
Costanzo V, Roviello GN (2023) “The potential role of vaccines in preventing Antimicrobial Resistance (AMR): an update and future perspectives,” pp. 1–21,
Ács N, Gambino M, Brøndsted L (October, 2020) Bacteriophage enumeration and detection methods. Front Microbiol 11. https://doi.org/10.3389/fmicb.2020.594868
Helmy YA et al (2023) “Antimicrobial Resistance and recent Alternatives to Antibiotics for the control of bacterial pathogens with an emphasis on Foodborne Pathogens,”
Pimenta J, Pinto AR, Jos M (2023) “Equine gram-negative oral microbiota: an Antimicrobial Resistances Watcher ?,” pp. 1–11,
Islam S (2023) “A comprehensive review on bacterial vaccines combating Antimicrobial Resistance in Poultry,”
Łobocka M, Dąbrowska K, Górski A (2021) Engineered Bacteriophage therapeutics: Rationale, Challenges and Future. BioDrugs 35(3):255–280. https://doi.org/10.1007/s40259-021-00480-z
Liu R et al (2022) Bacteriophage therapy in aquaculture: current status and future challenges. Folia Microbiol (Praha). https://doi.org/10.1007/s12223-022-00965-6
Choi Y, Ha E, Kong M, Ryu S (2023) A novel chimeric endolysin with enhanced lytic and binding activity against Clostridium perfringens. LWT 181:114776. https://doi.org/10.1016/j.lwt.2023.114776
Ali S, Karaynir A, Salih H, Oncü S, Bozdo B (2023) Characterization, genome analysis and antibiofilm efficacy of lytic Proteus phages RP6 and RP7 isolated from university hospital sewage. vol 326 no October 2022. https://doi.org/10.1016/j.virusres.2023.199049
Release JN (2019) “New report calls for urgent action to avert antimicrobial resistance crisis,”
Franco EG (2020) “Global Risks : An Unsettled World,” Glob. Risks Rep, pp. 8–17, 2020
Dedrick RM et al (2019) Disseminated drug resistant Mycobacterium abscessus. 25(5):730–733. https://doi.org/10.1038/s41591-019-0437-z.Engineered
Collaborators AR (2022) Articles Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. 6736(21). https://doi.org/10.1016/S0140-6736(21)02724-0
Infections B (2023) “Current Promising Strategies against Antibiotic-Resistant Bacterial Infections,”
Jeje O, Ewunkem AJ, Jeffers-francis LK (2023) “Serving two masters: Effect of Escherichia coli Dual Resistance on Antibiotic susceptibility,” pp. 1–20,
Morris C, Wickramasingha D, Abdelfattah EM, Pereira RV, Okello E, Maier G (2023) Prevalence of antimicrobial resistance in fecal Escherichia coli and Enterococcus spp. isolates from beef cow-calf operations in northern California and associations with farm practices. no Febr 1–14. https://doi.org/10.3389/fmicb.2023.1086203
Cristiane R, Leite T, Antônio T, Mendes DO (2023) Potential of the endogenous and artificially inserted CRISPR-Cas system for controlling virulence and antimicrobial resistance of food pathogens. Food Chem Adv 2:100229. no. February10.1016/j.focha.2023.100229
Gahamanyi N, Umuhoza T, Saeed SI, Mayigane LN, Hakizimana JN (2023) “A review of the important Weapons against Antimicrobial Resistance in Sub-Saharan Africa,” pp. 136–156,
Hyman P (2019) Phages for phage therapy: isolation, characterization, and host range breadth. Pharmaceuticals 12(1). https://doi.org/10.3390/ph12010035
Dedrick RM et al (2019) Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat Med 25(5):730–733. https://doi.org/10.1038/s41591-019-0437-z
Liang S et al (2023) “Bacteriophage therapy as an application for bacterial infection in China,” pp. 1–20,
Mahler M, Costa AR, Brouns SJJ, Van Beljouw SPB, Fineran PC (2023) Trends in Biotechnology Approaches for bacteriophage genome engineering. Trends Biotechnol 41(5):669–685. https://doi.org/10.1016/j.tibtech.2022.08.008
Sun Q et al (2023) Advance on Engineering of Bacteriophages by Advance on Engineering of Bacteriophages by Synthetic Biology. https://doi.org/10.2147/IDR.S402962
Kim K, Shin J, Kang TA, Kim B, Kim WC (2023) CRISPR/Cas9-mediated AtGATA25 mutant represents a novel model for regulating hypocotyl elongation in Arabidopsis thaliana. Mol Biol Rep 50(1):31–41. https://doi.org/10.1007/s11033-022-07926-9
Mathew SM (2023) Strategies for generation of mice via CRISPR/HDR-mediated knock-in. Mol Biol Rep 50(4):3189–3204. https://doi.org/10.1007/s11033-023-08278-8
Yang Y, Wang D, Lü P, Ma S, Chen K (2023) Research progress on nucleic acid detection and genome editing of CRISPR/Cas12 system. Mol Biol Rep 50(4):3723–3738. https://doi.org/10.1007/s11033-023-08240-8
Cameron P et al (2019) Harnessing type I CRISPR–Cas systems for genome engineering in human cells. Nat Biotechnol 37(12):1471–1477. https://doi.org/10.1038/s41587-019-0310-0
Lenneman BR, Fernbach J, Loessner MJ, Lu TK, Kilcher S (2021) Enhancing phage therapy through synthetic biology and genome engineering. Curr Opin Biotechnol 68:151–159. https://doi.org/10.1016/j.copbio.2020.11.003
Wetzel KS et al (2021) CRISPY-BRED and CRISPY-BRIP: efficient bacteriophage engineering. Sci Rep 11(1):1–6. https://doi.org/10.1038/s41598-021-86112-6
Ramirez-Chamorro L, Boulanger P, Rossier O (2021) Strategies for bacteriophage T5 mutagenesis: expanding the Toolbox for Phage Genome Engineering. Front Microbiol 12. https://doi.org/10.3389/fmicb.2021.667332
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8(11):2281–2308. https://doi.org/10.1038/nprot.2013.143
States U et al (2022) “Molecular Epidemiology of Carbapenem-Resistant,” vol. 28, no. 6, https://doi.org/10.1089/mdr.2021.0352
Mu AR (2022) Analysis of Antibiotic Resistance and Biofilm-Forming capacity in tetracycline-resistant Bacteria from a Coastal lagoon. 28(6):654–659. https://doi.org/10.1089/mdr.2021.0255
Kang K, Imamovic L, Misiakou M, Bornakke M, Heshiki Y (2021) Expansion and persistence of antibiotic-specific resistance genes following antibiotic treatment. Gut Microbes 13(1):1–19. https://doi.org/10.1080/19490976.2021.1900995
Ndagi U, Falaki AA, Abdullahi M, Lawal M (2020) Antibiotic resistance: bioinformatics-based understanding as a functional strategy for drug. 18451–18468. https://doi.org/10.1039/d0ra01484b
Chen Y, Li W, Shi K, Fang Z, Yang Y, Zhang R (2023) “Isolation and characterization of a novel phage belonging to a new genus against Vibrio parahaemolyticus,” pp. 1–11,
Manohar P, Madurantakam Royam M, Loh B, Bozdogan B, Nachimuthu R, Leptihn S (2022) Synergistic Effects of phage-antibiotic combinations against Citrobacter amalonaticus. ACS Infect Dis 8(1):59–65. https://doi.org/10.1021/acsinfecdis.1c00117
Saini S, Shanbhag C, Saraogi I (2023) Chemical Approaches to tackle the Silent Pandemic of Antibiotic Resistance. no April. https://doi.org/10.51167/acm00051
Van Der Horst MA, Schuurmans JM, Smid MC (2011) De Novo Acquisition of Resistance to three antibiotics by Escherichia coli. 17(2). https://doi.org/10.1089/mdr.2010.0101
Ji X et al (2022) Molecular characteristics of extended-spectrum beta-lactamase-producing Escherichia coli strains. 28(6):750–757. https://doi.org/10.1089/mdr.2021.0298
Uba AI, Abdulazeez MA, Usman SS, Tabakoglu HO, Abubakar H (2015) “Phage Dısplay Technology As A Strong Alternatıve To Hybrıdoma Technology For Monoclonal Antıbody Productıon,”
Carroll D (2014) Genome engineering with targetable nucleases. Annu Rev Biochem 83:409–439. https://doi.org/10.1146/annurev-biochem-060713-035418
Blazanin M, Lam WT, Vasen E, Chan BK, Turner PE (2022) “Decay and damage of therapeutic phage OMKO1 by environmental stressors,” PLoS One, vol. 17, no. 2 February, pp. 1–13, https://doi.org/10.1371/journal.pone.0263887
Vera-Mansilla J, Sánchez P, Silva-Valenzuela CA, Molina-Quiroz RC (2022) Isolation and characterization of Novel Lytic Phages infecting Multidrug-Resistant Escherichia coli. Microbiol Spectr 10(1). https://doi.org/10.1128/spectrum.01678-21
Dong Y et al (2022) Characterization and genomic analysis of the First Podophage Infecting Shewanella, representing a novel viral cluster. Front Microbiol 13:1–13. https://doi.org/10.3389/fmicb.2022.853973
Majdani R, Ghahfarokhi ES (2022) Isolation and characterization of lytic bacteriophages against Pseudomonas aeruginosa isolates from human infections in the north-west of Iran. 14(2):203–213
Zelcbuch L et al (2021) Luminescent phage-based detection of klebsiella pneumoniae: from engineering to diagnostics. Pharmaceuticals 14(4). https://doi.org/10.3390/ph14040347
Pulkkinen EM, Hinkley TC, Nugen SR (2019) Utilizing in vitro DNA assembly to engineer a synthetic T7 nanoluc reporter phage for Escherichia coli detection. Integr Biol 11(3):63–68. https://doi.org/10.1093/intbio/zyz005
Braun P, Raab R, Bugert JJ, Braun S “Recombinant Reporter Phage rTUN1::,” 2023, https://doi.org/10.1021/acssensors.2c01822
Vogele K, Falgenhauer E, Von Schönberg S, Simmel FC, Pirzer T (2021) Small antisense DNA-Based gene silencing enables cell-free bacteriophage manipulation and genome replication. ACS Synth Biol 10(3):459–465. https://doi.org/10.1021/acssynbio.0c00402
Li M, Shi D, Li Y, Xiao Y, Chen M, Chen L (2020) Recombination of T4-like phages and its activity against pathogenic Escherichia coli in Planktonic and Biofilm Forms. Virol Sin 35(5):651–661. https://doi.org/10.1007/s12250-020-00233-2
Amankwah S, Abdella K, Kassa T (2021) Bacterial biofilm destruction: a focused review on the recent use of phage-based strategies with other antibiofilm agents. Nanotechnol Sci Appl 14:161–177. https://doi.org/10.2147/NSA.S325594
Mohammadi M, Saffari M, Siadat SD, Hejazi SH (2023) Isolation, characterization, therapeutic potency, and genomic analysis of a novel bacteriophage vB _ KshKPC – M against carbapenemase – producing Klebsiella pneumoniae strains (CRKP) isolated from ventilator – associated pneumoniae (VAP) infection of COVID – 19 patients. Ann Clin Microbiol Antimicrob. https://doi.org/10.1186/s12941-023-00567-1
Nobrega FL et al (2018) Targeting mechanisms of tailed bacteriophages. Nat Rev Microbiol 16:760
Lammens EM, Nikel PI, Lavigne R (2020) Exploring the synthetic biology potential of bacteriophages for engineering non-model bacteria. Nat Commun 11(1):1–14. https://doi.org/10.1038/s41467-020-19124-x
Isaev A, Andriianov A, Znobishcheva E, Zorin E, Morozova N, Severinov K (2022) Editing of phage genomes—recombineering-assisted SpCas9 modification of Model Coliphages T7, T5, and T3. Mol Biol 56(6):801–815. https://doi.org/10.1134/S0026893322060073
Mahler M, Costa AR, van Beljouw SPB, Fineran PC, Brouns SJJ (2022) “Approaches for bacteriophage genome engineering,” Trends Biotechnol, vol. xx, no. xx, pp. 1–17, https://doi.org/10.1016/j.tibtech.2022.08.008
Shen J, Zhou J, Chen G-Q, Xiu Z-L (2018) Efficient Genome Engineering of a virulent Klebsiella. J Virol 92(17):1–20
Hoshiga F, Yoshizaki K, Takao N, Miyanaga K, Tanji Y (2019) Modification of T2 phage infectivity toward Escherichia coli O157:H7 via using CRISPR/Cas9. FEMS Microbiol Lett 366(4):1–7. https://doi.org/10.1093/femsle/fnz041
Salmonella M, Almoghrabi SZ, El-obeid T, Al-hadidi SH, Al H (2022) Retail chicken carcasses as a Reservoir. 28(7):824–831. https://doi.org/10.1089/mdr.2021.0414
Duong MM, Carmody CM, Ma Q, Peters JE, Nugen SR (2020) Optimization of T4 phage engineering via CRISPR/Cas9. Sci Rep 10(1):1–9. https://doi.org/10.1038/s41598-020-75426-6
Xia X (2023) “Optimizing protein production in therapeutic phages against a bacterial Pathogen, Mycobacterium abscessus,” no. 1, pp. 189–209,
Palma M (2023) “Aspects of phage-based vaccines for protein and Epitope Immunization,” pp. 1–23,
Maffei E et al (2021) Systematic exploration of Escherichia coli phage-host interactions with the BASEL phage collection. PLoS Biol 19(11):e3001424. https://doi.org/10.1371/journal.pbio.3001424
Bayat F, Didar TF, Hosseinidoust Z (2021) Emerging investigator series: bacteriophages as nano engineering tools for quality monitoring and pathogen detection in water and wastewater. Environ Sci Nano 8(2):367–389. https://doi.org/10.1039/d0en00962h
Airola C et al (2023)
Bondad-reantaso MG et al (2022) “Review of alternatives to antibiotic use in aquaculture,” no. pp. 1–31, 2023, https://doi.org/10.1111/raq.12786
Pan P et al (2023) Bacteriophage – based techniques for elucidating the function of zebrafish gut microbiota. 2039–2059. https://doi.org/10.1007/s00253-023-12439-x
Acknowledgements
No acknowledgements.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Sani Sharif Usman and Evangeline Christina conceived the idea; Sani Sharif Usman designed the review and gathered the information; Sani Sharif Usman, Abdullahi Ibrahim Uba and Evangeline Christina wrote the manuscript. The authors read and approved the final manuscript as well as agreed to authorship and submission of the manuscript for review.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Authors have declared that no competing interest exists.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Usman, S.S., Uba, A.I. & Christina, E. Bacteriophage genome engineering for phage therapy to combat bacterial antimicrobial resistance as an alternative to antibiotics. Mol Biol Rep 50, 7055–7067 (2023). https://doi.org/10.1007/s11033-023-08557-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08557-4