Abstract
Aim
The current study aimed to evaluate the effects of caspase-8 (CASP8) and mitogen-activated protein kinase 1 (MAPK1) gene expression levels and their products on preventing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection.
Methods
A total of 40 patients (men, 15 [37.5%]; women, 25 [62.5%]) with COVID-19 infection were included in the current study. The patients were divided into four main groups based on disease severity: mild (n = 7), moderate (n = 10), severe (n = 14), and critical (n = 9). Individuals aged < 18 years and pregnant women were excluded. Patients were classified according to the World Health Organization (WHO) classification system (WHO/2019-nCoV/clinical/2021.1).
Results
Considering all groups, statistically significant differences were detected among all groups for both CASP82−ΔΔCt (p = 0.006) and MAPK1 2−ΔΔCt values (p = 0.015). Moreover, statistically significant differences were detected between mild and moderate (p = 0.013), moderate and critical (p = 0.018), and severe and critical (p = 0.023) groups for lymphocytes.
Conclusion
The CASP8/MAPK1 expression levels and/or its products are essential in preventing injury caused by COVID-19 infection. They play crucial roles in maintaining cellular homeostasis and viability. Furthermore, CASP8/MAPK1 levels can provide information about disease severity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The disease was named “coronavirus disease 2019 (COVID-19)” by the World Health Organization (WHO) in February after it was understood that the epidemic, which started with pneumonia of unknown etiology, was caused by a new coronavirus-related infection and associated with a seafood market in Wuhan, China, in December 2019 [1]. Coronaviruses are RNA viruses with rod-like protrusions on their surface and are enclosed and single-stranded and have positive polarity. They are called coronaviruses because the protrusions resemble “corona,” the Latin for “crown.” Coronaviruses belong to the Coronaviridae family, Orthocoronavirinae subfamily, which is divided into four genera: alpha, beta, gamma, and delta coronavirus [2].
Mitogen-activated protein kinases (MAPKs) may alter host defense and apoptotic mechanisms; they are a family of highly conserved serine-threonine protein kinases that link external stimuli, such as viral infections, to the cell nucleus and activate the MAPK pathway in viral infections [3]. In virus-infected cells, the MAPK cascade is also implicated in controlling immune response and apoptosis [4]. Various DNA and RNA viruses use MAPK cascades to activate signal transduction pathways in infected host cells. MAPK signaling can act as a positive or negative viral replication regulator. MAPK pathways can be triggered by the presence of both active and dormant viral cells [3].
CASP8 (aspartate-specific cysteine protease), has roles in the induction of exogenous apoptosis, the inhibition of necroptosis, and is a critical regulator of programmed cell death activation [5] . CASP8 can control inflammasome activation and generation and maturation of pro-inflammatory cytokines IL1 and IL18 and regulate inflammation withouth death receptor signaling [5].
CASP8 and MAPK1 have been studied in the context of viral infections [6]. In addition to revealing the roles of genes in the etiopathogenesis of diseases [7, 8], the detection of both the expression levels of the genes [9, 10] and their products [11,12,13,14] are crucial to understand their function and their role maintaining cellular homeostasis and viability.The current study aimed to evaluate the effects of CASP8 and MAPK1 gene expression levels and their products to determine their function in individuals with different severities of COVID-19 infection.
Methods
Patients and groups
A total of 40 patients (men, 15 [37.5%]; women, 25 [62.5%]) with COVID-19 infection were included in the current study. The patients were divided into four main groups: mild disease (n = 7), moderate (n = 10), severe (n = 14) and critical (n = 9). Patients aged under 18 years of age and pregnant women were excluded.
Mild disease: Individuals who have any of the various signs and symptoms of COVID-19 (e.g., fever, cough, sore throat, malaise, headache, muscle pain, nausea, vomiting, diarrhea, and loss of taste and smell) but do not have shortness of breath, dyspnea, or abnormal chest imaging.
Moderate disease: Individuals who show evidence of lower respiratory disease during clinical assessment or imaging and have an oxygen saturation (SpO2) of ≥ 94% on room air at sea level.
Severe disease: Individuals who have SpO2 of < 94% on room air at sea level, ratio of arterial partial pressure of oxygen to fraction of inspired oxygen (PaO2/FiO2) < 300 mmHg, respiratory frequency > 30 breaths/min, or lung infiltrates > 50%.
Critical disease: Individuals who have respiratory failure, septic shock, and/or multiple organ dysfunction (https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/).
Ethics
The study was approved by the Ministry of Health and Local Ethics Committee (Karabuk University Local Ethic Committee’s approval document dated November 11, 2020, and numbered 338). Individuals with COVID-19 infection were divided into four main groups based on disease severity: mild (n = 7), moderate (n = 10), severe (n = 14), and critical (n = 9). Patients were classified according to the WHO classification system (WHO/2019-nCoV/clinical/2021.1). Individuals aged < 18 years and pregnant women were excluded from the study.
Detection of blood parameters
The Beckman Coulter LH 780 analyzer was used to examine the values of hemogram parameters (hemoglobin [HB], platelet, and lymphocyte). Using original commercial kits, the Roche Cobas Integra 400 plus auto-analyzer was used to assess C-reactive protein (CRP) readings.
The Cobas Roche t511 analyzer was used to measure D-dimer levels. The serum lactate dehydrogenase (LDH) and ferritin levels were measured using a Roche Cobas 702 auto-analyzer.
Polymerase chain reactions were performed using SARS-CoV-2 RTqPCR Detection Kit (Bioeksen, Turkey) via a real-time polymerase chain reaction (PCR) analyzer (Anatolia Geneworks, Turkey).
Detection of human CASP8 and MAPK1 levels via enzyme-linked immunosorbent assay (ELISA)
Both human CASP8 and MAPK1 levels were determined from blood serum samples of patients using the human CASP8 (Invitrogen) (Catalog No. BMS2024) and MAPK1 (Fine Test) (Catalog No. EH0665) kits via the ELISA method based on biotin double antibody sandwich technology. Bio-Tek Instruments E.L.X. 50 Strip Washer was used as a microplate washer. Each well’s optical density was determined immediately using a 450-nm wavelength microplate reader (Bio-Tek Instruments E.L.X. 800 Absorbance Microplate Reader). The findings of the samples were obtained by substituting the device’s absorbance values in the calibration graph.
RNA isolation and cDNA synthesis
The patient’s peripheral blood samples for RNA isolation were placed in RNase and DNase-free tubes containing ethylenediaminetetraacetic acid. Then, according to the manufacturer’s instructions, RNA was extracted from peripheral blood samples of patients using RiboEx (Catalog No. 301-001) and Hybrid-R (Catalog No. 305-101) kits. Measurements were obtained with a spectrophotometer to determine the adequacy of the RNA quality for the investigation. The extracted RNA was kept in RNase-free water at − 20 °C. Using the ABT cDsNA Synthesis Kit (High Capacity) (Catalog No. C03-01-05), cDNA was obtained from isolated RNA Mastermix used for cDNA synthesis including 2 µL RNA, 2 µL 10X RTase Reaction Buffer, 1 µL dNTP mix, 3.5 µL RNase-free distilled water, 1 µL Reverse Transcriptase, 0.5 µL RNase Inhibitor, and 2 µL Random Hexamer.
Relative gene expressions of CASP8 and MAPK1 gene by real-time qPCR
The expression levels of CASP8, MAPK1, and the reference gene (ACTB) were detected via the Applied Biosystems 7500 and ViiA7 real-time PCR systems for each cDNA sample of the patients. The ABT 2X qPCR SYBR-Green Mastermix (without ROX) (Catalog No. Q03-01-05) kit was used for the PCR in a final volume of 20 µL: 4 µL cDNA, 10 µL 2X MasterMix (with SYBR-Green), 3 µL RNase-free distilled water, 1 µL QN ROX Reference Dye (Applied Biosystems cycler only), 1 µL forward primer (10 µM), and 1 µL reverse primer (10 µM). Cycle conditions of the real-time qPCR were initial denaturation at 95 °C for 300 s, 40 cycles of denaturation at 95 °C for 15 s, and 40 cycles of annealing at 60 °C for 60 s. ACTB transcript was used as a reference for the quantitation of mRNA expressions and normalized according to the control group. Calculation of fold change had been calculated via processing ΔΔCt values as 2−ΔΔCt.
Statistical analysis
The data were analyzed via the Statistical Package for Social Sciences (IBM Corp., Armonk, NY, USA) for Windows 23.0. The Shapiro-Wilk test was used in determining the data distribution. Because the data were not normally distributed (p < 0.05), nonparametric tests were used for statistical analysis. In addition to descriptive statistics (number, mean, standard deviation [SD]) for each variable, the Kruskal-Wallis test was used to compare all groups; additionally, a polynomial regression test was performed. A p-value of < 0.05 was accepted as statistically significant.
Results
The mean age of patients was 63.225 ± 14.995 years. For age, no statistically significant differences were found among all groups (2 = 1.569, p = 0.666) (Table 1).
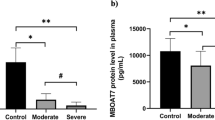
The mean CASP8 2−ΔΔCt and MAPK1 2−ΔΔCt expression and CASP8 and MAPK1 serum levels of each group are presented in Table 1; Fig. 1b and c, and Fig. 2b and c. When all the groups were compared, statistically significant differences were detected among all groups for both CASP8 2−ΔΔCt (χ2 = 12.413; p = 0.006) (Table 1) (Fig. 1b) and MAPK1 2−ΔΔCt values (χ2 = 10.429; p = 0.015) (Table 1) (Fig. 2b). To detect this differences caused from which groups, binary comparison of the groups was performed. Statistically significant differences were found between mild and severe groups (Z = − 2.164; p = 0.030), between mild and critical groups (Z = − 2.17; p = 0.030), between moderate and severe groups (Z = − 2.284; p = 0.022), and between moderate and critical groups (Z = − 2.613; p = 0.009) for CASP8 2−ΔΔCt values. However, there were no differences between mild and moderate groups (Z = − 1.464; p = 0.143) and between severe and critical groups (Z = − 1.512; p = 0.131) (Table 2). The differences between the mild and critical groups (Z = − 2.276; p = 0.023) and between the moderate and critical groups (Z = − 1.922; p = 0.05) were statistically significant in terms of serum CASP8 level (Table 2).
Demonstrative examples of COVID-19 infection. The patients were divided into four main groups based on the severity of COVID-19 infection: mild, moderate, severe, and critical. Both CASP8 2−ΔΔCt and CASP8 levels of patients were detected. The COVID-19 infection causes cellular damage, and a response occurs in the nucleus of cells. The CASP8 gene is transcribed CASP8 mRNA, and CASP8 proteins are translated from mature CASP8 mRNA. The ELISA standard curve is presented in Fig. 1(a). Statistically significant differences were detected among all groups for CASP8 2−ΔΔCt levels. As the severity of COVID-19 infection increased, the expression level of CASP8 gene significantly decreased. Considering the CASP8 gene expression level in the pairwise comparison, there were statistically significant differences between mild and severe, mild and critical, moderate and severe, moderate and critical groups [Fig. 1(b)]. Serum caspase8 levels of groups are shown in Fig. 1(c). As the severity of COVID-19 infection increased, the serum CASP8 levels significantly decreased. There was a statistically significant difference between mild and critical and between moderate and critical groups in terms of serum CASP8 levels
Demonstrative examples of COVID-19 infection. The patients were divided into four main groups based on the severity of COVID-19 infection: mild, moderate, severe, and critical. Both MAPK1 2−ΔΔCt and MAPK1 levels of patients were detected. The COVID-19 infection causes cellular damage, and a response occurs in the nucleus of cells. MAPK1 gene is transcribed MAPK1mRNA, and MAPK1 proteins are translated from mature MAPK1 mRNA. The ELISA standard curve is presented in Fig. 2(a). Statistically significant differences were detected among all groups for MAPK1 2−ΔΔCt levels. As the severity of COVID-19 infection increased, the expression level of the MAPK1 gene significantly decreased. Considering the MAPK1 gene expression level in a pairwise comparison, there was a statistically significant difference between mild and critical and between moderate and critical groups [Fig. 2(b)]. Serum MAPK1 levels of groups are presented in Fig. 2(c). When the severity of COVID-19 infection increased, the serum level significantly decreased. Considering the serum MAPK1 levels in pairwise comparison, there was no statistically significant differences between the groups
When the binary comparison of the groups was performed based on MAPK1 2−ΔΔCt, statistically significant differences were found between the mild and critical groups (Z = − 2.170; p = 0.030), moderate and severe groups (Z = − 2.197; p = 0.028), and moderate and critical groups (Z = − 2.531; p = 0.011). When the serum MAPK1 levels of all groups were evaluated, there were no statistically significant differences between them (χ2 = 3.249; p = 0.355) (Table 2).
In Table 1, the hemogram values of each group are listed. When all groups were evaluated, statistically significant differences in D-dimer (p = 0.030) and lymphocyte (p = 0.009) levels were found (Table 1) (Fig. 3). When binary comparison of the groups was performed to determine which groups were responsible for the differences, statistically significant differences were found between the mild and severe groups (Z = − 2.164, p = 0.030) and between mild and critical groups (Z = − 2.911, p = 0.004) for D-dimer levels (Table 2) (Fig. 3).
As the severity of COVID-19 infection increased, D-dimer level significantly increased. Considering the D-dimer level in the pairwise comparison, there was a statistically significant difference between mild and critical and between mild and severe groups (a) and lymphocytes (b). Considering the lymphocyte level in the pairwise comparison, there was a statistically significant difference between mild and moderate, moderate and critical, and severe and critical groups
The differences between mild and moderate (Z = − 2.490; p = 0.013), moderate and critical (Z = − 2.370; p = 0.018), and severe and critical (Z = − 2.269, p = 0.023) groups were also statistically significant for lymphocytes (Table 2) (Fig. 3).
Discussion
This study found a statistically significant difference for both CASP8 2−ΔΔCt, and MAPK1 2−ΔΔCt values among all patient groups. Moreover, CASP8 levels are inversely related to disease severity in all study population groups, but the difference was close to statistically significant among them. Considering the serum CASP8 levels, as the severity of COVID-19 infection increased, the serum CASP8 levels significantly decreased. There was a statistically significant difference between mild and critical and between moderate and critical groups in terms of serum CASP8 levels. Although there was a statistically significant difference between mild and critical and between moderate and critical groups in terms of CASP8 2−ΔΔCt, CASP8, and MAPK1 2−ΔΔCt levels. Conversely, no difference was detected in MAPK1 level between the whole study population. Furthermore, we found a significant difference between the groups for D-dimer and lymphocyte levels, which are among the poor prognostic factors (LDH, ferritin, D-dimer, hemoglobin, lymphocyte, CRP) and used clinically for COVID-19 infection.
When the HB [15] and lymphocyte [16] levels decrease, LDH [17], D-dimer [18], and ferritin [19] levels and disease severity increase. Similarly, we found that the disease severity increased as the D-dimer level, one of the poor prognostic factors used in the clinic, increased. Lymphopenia was detected as the disease severity increased. The main reason we could not find a statistical significance for CRP and ferritin levels, which are the indicators of inflammation, may be caused the use of anti-inflammatory medications, such as prednisolone and IL-1 and IL-6 antagonists, by severe and critical patients.
Because MAPKs are heterogeneous kinases that can phosphorylate serine and threonine residues in various proteins, the formation of MAPK-associated pathways is thought to be the most likely cause of significant pro-inflammatory cytokine production (activation of the host’s innate response) [20, 21].
Tiwari et al. designed an experimental study to describe the viral pathogenesis mechanism associated with complement and coagulation pathways in COVID-19 infection [21]. As a result, they hypothesized that the MAPK signaling pathway is essential for the innate immune response to COVID-19 infection and the pathophysiology that underpins it [21]. According to our results, as the severity of COVID-19 infection increased, the expression level of the MAPK1 gene significantly decreased. Considering the MAPK1 gene expression level in pairwise comparison, there was a statistically significant difference between mild and critical and between moderate and critical groups. Additionally, when the severity of COVID-19 infection increased, although the serum MAPK1 levels decreased, there were no statistically significant differences in pairwise comparison of the groups.
Patients with COVID-19 infections have a severe inflammatory reaction to the virus and various systemic symptoms linked to the release of high amounts of pro-inflammatory cytokines (cytokine storms) [22]. Pyroptosis and necroptosis are inflammatory cell death methods that release cytokines and other cellular components to trigger inflammation and alert immune cells to pathogenic or sterile damage, whereas apoptosis is considered immune-silent [22]. CASP3 and CASP7 promote apoptosis following the activation of upstream initiator CASP8/CASP10 or CASP9 [23].
The MAPK pathway, which regulates the expression of cytokines and chemokines, promotes the formation of cytokine storms. The type I interferon, MAPK, and ferroptosis pathways were activated while the disease was active, and they gradually recovered as the patient’s health improved [24].
In the apoptosis process, CASP8 plays the initiator role, whereas CASP3 and CASP6 play the executioner role. In lung epithelial cells, COVID-19 infection activates CASP8, resulting in cell death and inflammatory cytokine processing. The analysis of postmortem lung sections of fatal COVID-19 patients revealed not only massive inflammatory cell infiltration, necrotic cell debris, and pulmonary interstitial fibrosis but also apoptosis and necroptosis [25]. Gulhan et al. [9] investigated the relationship between CASP3 and severity of COVID-19 infection and discovered a significant difference in CASP3 2−ΔΔCt levels among all groups in a disease severity classification. They found that statistically significant differences between mild and critical groups and between intermediate and critical groups for CASP3 2−ΔΔCt levels. Statistically significant changes in CASP3 levels between mild and severe groups and between mild and critical groups were also detected [24].
We concluded that, when the CASP8 2−ΔΔCt, MAPK1 2−ΔΔCt, and CASP8 levels decreased, the disease severity increased. Moreover, we detected a significant relationship between both CASP8 2−ΔΔCt and MAPK1 2−ΔΔCt levels and disease severity. CASP8 and MAPK1 gene expression levels may provide information about the severity of COVID-19 infection. CASP8 and MAPK1 gene expression begins to increase when the virus is encountered. The severity of the disease probably decreases when the CASP8 and MAPK1 gene expression capacity increase.
This study would have more impact if a control group of healthy individuals could have been included and then compared to the different SARS-CoV-2 groups of patients. This is a limitation of the study, and including a control group is still needed in a future study.
CASP8 and MAPK1 gene expression and/or their products may change depending on the cellular damage caused by COVID-19 infection. This cycle can be repeated until the system reaches equilibrium. CASP8 and MAPK1 expression levels should not be lower than those required to trigger and maintain apoptosis and should not be adequately high to cause a cytokine storm. This should be at a level that keeps the body in a state of homeostasis. Therefore, it is logical to say that the CASP8/MAPK gene and/or its products play a role in preventing damage and maintaining cellular homeostasis and viability. Furthermore, CASP8/MAPK level can serve as an indicator of COVID-19 disease severity. The discovery of disease pathology processes during COVID-19 infection may pave the way for developing evidence-based treatment solutions to address this public health problem. More research on the activity levels of CASP8 and MAPK1 is needed to better understand the current condition.
References
Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W (2019) A novel coronavirus from patients with Pneumonia in China, 2019. N Engl J Med 382:727–733
Malik YA (2020) Properties of coronavirus and SARS-CoV-2. Malays J Pathol 42:3–11
Mohanta TK, Sharma N, Arina P, Defilippi P (2020) Molecular Insights into the MAPK Cascade during viral infection: potential crosstalk between H.C.Q. and H.C.Q. Analogues.Biomed Res Int. DOI: 10.1155/2020/8827752
Bian J, Wang K, Kong X, Liu H, Chen F, Hu M, Zhang X, Jiao X, Ge B, Wu Y, Meng S (2011) Caspase- and p38-MAPK-dependent induction of apoptosis in A549 lung cancer cells by newcastle disease virus. Arch Virol 156:1335–1344
Tummers B, Green DR (2017) Caspase-8: regulating life and death. Immunol Rev 277:76–89
Galganska H, Jarmuszkiewicz W, Galganski L (2021) Carbon dioxide inhibits COVID-19-type pro-inflammatory responses through extracellular signal-regulated kinases 1 and 2, novel carbon dioxide sensors. Cell Mol Life Sci 78:8229–8242
Türay S, Eröz R, Başak, (2021) AN A novel pathogenic variant in the 3’ end of the AGTPBP1 gene gives rise to neurodegeneration without cerebellar atrophy: an expansion of the disease phenotype? Neurogenetics 22(2):127–132. https://doi.org/10.1007/s10048-021-00643-8
Eroz R, Damar İH, Kılıçaslan O (2020) Thrombosis risk of Alport syndrome patients: evaluation of cardiological, clinical, biochemical, genetic and possible causes of inherited thrombophilia and identification of a novel COL4A3 variant. Blood Coagul Fibrinolysis 31(4):264–269. doi: https://doi.org/10.1097/MBC.0000000000000911
Yildiz Gulhan P, Eroz R, Ataoglu O, İnce N, Davran F, Öztürk CE, Gamsızkan Z, Balbay O (2022) The evaluation of both the expression and serum protein levels of Caspase-3 gene in patients with different degrees of SARS-CoV2 infection. J Med Virol 94(3):897–905. doi: https://doi.org/10.1002/jmv.27362
Tasdemir S, Eroz R, Dogan H, Erdem HB, Sahin I, Kara M, Engin RI, Turkez H (2016) Association between human hair loss and the expression levels of nucleolin, nucleophosmin, and UBTF genes. Genet Test Mol Biomark 20(4):197–202. https://doi.org/10.1089/gtmb.2015.0246
Kabaklıoğlu M, Eroz R, Kaya M (2021) May argyrophilic nucleolar organizer regions be used as a biomarker for the detection of the degree of ischemic damage instead of tunel in testicular torsion? Med (Kaunas) 57(11):1177. https://doi.org/10.3390/medicina57111177
Damar İH, Eroz R (2022) Argyrophilic nucleolar organizer regions as new biomarkers in ST-Elevation myocardial infarction. J Cardiovasc Dev Dis 9(2):58. doi: https://doi.org/10.3390/jcdd9020058
Yesildag K, Kokulu K, Mutlu H, Eroz R, Taha-Sert E, Saritas A (2021) Argyrophilic nucleolar organizer regions as a promising biomarker for the detection of brain hypoxia levels caused by different doses of carbon monoxide poisoning. Gac Med Mex 157(6):610–617. https://doi.org/10.2487/GMM.M21000625
Kaya M, Eroz R, Kabakliogli M (2022) Expression of nucleolin, nucleophosmin, upstream binding transcription factor genes and propolis in wound models. J Wound Care 31(Sup10):S28–S40. https://doi.org/10.1296/jowc.2022.31.Sup10.S28
Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS (2020) Clinical characteristics of Coronavirus disease 2019 in China. N Engl J Med 382:1708–1720
Tan L, Wang Q, Zhang D, Ding J, Huang Q, Tang YQ, Wang Q, Miao H, (2020) Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study [published correction appears in Signal Transduct Target Ther. (2020 Apr) 29;5(1):61]. Signal Transduct Target Ther 5(1):1–3
Niu Y, Zhan Z, Li J, Shui W, Wang C, Xing Y, Zhang C (2021) Development of a predictive model for mortality in hospitalized patients with COVID-19. Disaster Med Public Health Prep 8:1–9. doi: https://doi.org/10.1017/dmp.2021.8
Ozen M, Yilmaz A, Cakmak V, Beyoglu R, Oskay A, Seyit M, Senol H (2021) D-Dimer as a potential biomarker for disease severity in COVID-19. Am J Emerg Med 40:55–59
Deng F, Zhang L, Lyu L, Lu Z, Gao D, Ma X, Guo Y, Wang R, Gong S, Jiang W (2021) Increased ferritin levels on admission predict intensive care unit mortality in patients with COVID-19. Med Clin (Engl Ed) 156:324–331
Arthur JS, Ley SC (2013) Mitogen-activated protein kinases in innate immunity. Nat Rev Immunol 13:679–692
Tiwari R, Mishra AR, Mikaeloff F, Gupta S, Mirazimi A, Byrareddy SN, Neogi U, Nayak D (2020) In silico and in vitro studies reveal complement system drives coagulation cascade in SARS-CoV-2 pathogenesis. Comput Struct Biotechnol J 18:3734–3744
Karki R, Sharma BR, Tuladhar S, Williams EP, Zalduondo L, Samir P, Zheng M, Sundaram B, Banoth B, Malireddi RKS, Schreiner P, Neale G, Vogel P, Webby R, Jonsson CB, Kanneganti TD (2021) Synergism of TNF-α and IFN-γ triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell 184(1):149–168. https://doi.org/10.1016/j.cell.2020.11.025
akhani SA, Masud A, Kuida K, Porter GA Jr, Booth CJ, Mehal WZ, Inayat I, Flavell RA (2006) Caspases 3 and 7: key mediators of mitochondrial events of apoptosis. Science 311:847–851. doi: https://doi.org/10.1126/science.1115035
Huang L, Shi Y, Gong B, Jiang L, Zhang Z, Liu X, Yang J, He Y, Jiang Z, Zhong L, Tang J, You C, Jiang Q, Long B, Zeng T, Luo M, Zeng F, Zeng F, Wang S, Yang X, Yang Z (2021) Dynamic blood single-cell immune responses in patients with COVID-19. Signal Transduct Target Ther 6:110. doi: https://doi.org/10.1038/s41392-021-00526-2
Li S, Zhang Y, Guan Z, Li H, Ye M, Chen X, Shen J, Zhou Y, Shi ZL, Zhou P, Peng K (2020) SARS-CoV-2 triggers inflammatory responses and cell death through caspase-8 activation. Signal Transduct Target Ther 5:235. doi: https://doi.org/10.1038/s41392-020-00334-0
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interest
This study was supported by the Karabuk University coordinatorship of scientific research projects. (Project no: KBÜBAP-21-ABP-035)
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Acat, M., Yıldız Gülhan, P., Eröz, R. et al. Evaluation of both expression and serum protein levels of caspase-8 and mitogen-activated protein kinase 1 genes in patients with different severities of COVID-19 infection. Mol Biol Rep 50, 3241–3248 (2023). https://doi.org/10.1007/s11033-023-08244-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08244-4