Abstract
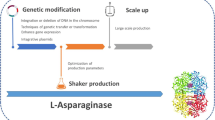
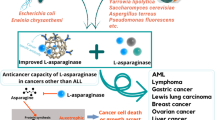
l-asparaginases are mostly obtained from bacterial sources for their application in the therapy and food industry. Bacterial l-asparaginases are employed in the treatment of Acute Lymphoblastic Leukemia (ALL) and its subtypes, a type of blood and bone marrow cancer that results in the overproduction of immature blood cells. It also plays a role in the food industry in reducing the acrylamide formed during baking, roasting, and frying starchy foods. This importance of the enzyme makes it to be of constant interest to the researchers to isolate novel sources. Presently l-asparaginases from E. coli native and PEGylated form, Dickeya chrysanthemi (Erwinia chrysanthemi) are in the treatment regime. In therapy, the intrinsic glutaminase activity of the enzyme is a major drawback as the patients in treatment experience side effects like fever, skin rashes, anaphylaxis, pancreatitis, steatosis in the liver, and many complications. Its significance in the food industry in mitigating acrylamide is also a major reason. Acrylamide, a potent carcinogen was formed when treating starchy foods at higher temperatures. Acrylamide content in food was analyzed and pre-treatment was considered a valuable option. Immobilization of the enzyme is an advancing and promising technique in the effective delivery of the enzyme than in free form. The concept of machine learning by employing the Artificial Network and Genetic Algorithm has paved the way to optimize the production of l-asparaginase from its sources. Gene-editing tools are gaining momentum in the study of several diseases and this review focuses on the CRISPR-Cas9 gene-editing tool in ALL.

Similar content being viewed by others
References
Oliveira M, Sampaio KC, Oliveira AC et al (2015) Outcome of children and adolescents with lymphoblastic lymphoma. Rev Assoc Med Bras 61(5):417–422. https://doi.org/10.1590/1806-9282.61.05.417
Sousa DWL, Ferreira FVA et al (2015) Acute lymphoblastic leukemia in children and adolescents: prognostic factors and analysis of survival. Rev Bras Hematol Hemoter 7:223–229. https://doi.org/10.1016/j.bjhh.2015.03.009
Arber DA, Orazi A, Hasserjian R et al (2016) The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 127:2391–2406. https://doi.org/10.1182/blood-2016-03-643544
VeskovićMoračanin SM, Dukić DA, Memiši NR (2014) Bacteriocins produced by lactic acid bacteria-a review. Acta Periodica Technologica 45:271–283. https://doi.org/10.2298/APT1445271V
Basso A, Serban S (2019) Industrial applications of immobilized enzymes-a review. Molecular Catalysis 479:110607. https://doi.org/10.1016/j.mcat.2019.110607
Noma SAA, Ulu A, Acet O, Sanz R, Sanz-Perez ES, Odabasi M, Ates B (2020) Comparative study of ASNase immobilization on tannic acid-modified magnetic Fe3O4/SBA-15 nanoparticles to enhance stability and reusability. New J Chem 44:4440–4451. https://doi.org/10.1039/D0NJ00127A
Agrawal S, Kango N (2019) Development and catalytic characterization of l-asparaginase nano-bioconjugates. Int J Biol Macromol 135:1142–1150. https://doi.org/10.1016/j.ijbiomac.2019.05.154
Ulu A (2020) Metal–organic frameworks (MOFs): a novel suort platform for ASNase immobilization. J Mater Sci 55:6130–6144. https://doi.org/10.1007/s10853-020-04452-6
Do TT, Do TP, Nguyen TN, Nguyen TC, Phuong V, Nguyen TGA (2019) Nanoliposomal l-asparaginase and its antitumor activities in Lewis lung carcinoma tumor-induced BALB/c mice. Adv Mat Sci Eng 2019:3534807. https://doi.org/10.1155/2019/3534807
Bahreini E, Aghaiypour K, Abbasalipourkabir R et al (2014) Preparation and nanoencapsulation of l-asparaginase II in chitosan-tripolyphosphate nanoparticles and in vitro release study. Nanoscale Res Lett 9:340. https://doi.org/10.1186/1556-276X-9-340
Kishore V, Nishita KP, Manonmani HK (2015) Cloning, expression and characterization of l-asparaginase from Pseudomonas fluorescens for large scale production in E. coli BL21. 3 Biotech 5(6):975–981. https://doi.org/10.1007/s13205-015-0300-y
Einsfeldt K, Baptista IC, Pereira JCCV, Costa-Amaral IC, Costa ESd, Ribeiro MCM et al (2016) Recombinant l-asparaginase from Zymomonas mobilis: a potential new antileukemic agent produced in Escherichia coli. PLoS ONE 11(6):e0156692. https://doi.org/10.1371/journal.pone.0156692
Husain I, Sharma A, Kumar S, Malik F (2016) Purification and characterization of glutaminase free asparaginase from Enterobacter cloacae: in-vitro evaluation of cytotoxic potential against human myeloid leukemia HL-60 cells. PLoS ONE 11(2):e0148877. https://doi.org/10.1371/journal.pone.0148877
Meghavarnam AK, Janakiraman S (2018) Evaluation of acrylamide reduction potential of l-asparaginase from Fusarium culmorum (ASP-87) in starchy products. LWT - Food Sci Technol 89:32–37. https://doi.org/10.1016/j.lwt.2017.09.048
Ganeshan S, Birendranarayan A et al (2016) Hemocompatible glutaminase free l-asparaginase from marine Bacillus tequilensis PV9W with anticancer potential modulating p53 expression. RSC Adv 6:25943–25951. https://doi.org/10.1039/C6RA00727A
Shi R, Liu Y, Mu Q, Jiang Z, Yang S (2017) Biochemical characterization of a novel l-asparaginase from Paenibacillus barengoltzii being suitable for acrylamide reduction in potato chips and mooncakes. Int J Bio Macromol 96:93–99. https://doi.org/10.1016/j.ijbiomac.2016.11.115
Zuo S, Zhang T, Jiang B, Mu W (2015) Reduction of acrylamide level through blanching with treatment by an extremely thermostable l-asparaginase during French fries processing. Extremophiles 19:841–851. https://doi.org/10.1007/s00792-015-0763-0
Radha R, Arumugam N, Gummadi SN (2018) Glutaminase free l-asparaginase from vibrio cholerae: heterologous expression, purification and biochemical characterization. Int J Biol Macromol 111:129–138. https://doi.org/10.1016/j.ijbiomac.2017.12.165
Sun Z, Qin R, Li D, Ji K, Wang T, Cui Z, Huang Y (2016) A novel bacterial type II l-asparaginase and evaluation of its enzymatic acrylamide reduction in French fries. Int J Biol Macromol 92:232–239. https://doi.org/10.1016/j.ijbiomac.2016.07.031
Nikolaos E, Labrou MM, Muharram, (2016) Biochemical characterization and immobilization of Erwinia carotovora l-asparaginase in a microplate for high-throughput biosensing of l-asparagine. Enzyme Microb Technol 92:86–93. https://doi.org/10.1016/j.enzmictec.2016.06.013
Ulu A, Koytepe S, Ates B (2016) Synthesis and characterization of biodegradable pHEMA-starch composites for immobilization of l-asparaginase. Polym Bull 73:1891–1907. https://doi.org/10.1007/s00289-015-1583-1
Ulu A, Koytepe S, Ates B (2016) Synthesis and characterization of PMMA composites activated with starch for immobilization of l-asparaginase. J Appl Polym Sci 133:43421. https://doi.org/10.1002/a.43421
Ulu A, Ozcan I, Koytepe S, Ates B (2018) Design of epoxy-functionalized Fe3O4@MCM-41 core-shell nanoparticles for enzyme immobilization. Int J Biol Macromol 115:1122–1130. https://doi.org/10.1016/j.ijbiomac.2018.04.157
Ashok A, Devarai SK (2019) l-asparaginase production in rotating bed reactor from Rhizopus microspores IBBL-2 using immobilized Ca-alginate beads. 3 Biotech 9(9):1–10. https://doi.org/10.1007/s13205-019-1883-5
Alam S, Ahmad R, Pranaw K, Mishra P, Khare SK (2018) Asparaginase conjugated magnetic nanoparticles used for reducing acrylamide formation in food model system. Bioresour Technol 269:121–126. https://doi.org/10.1016/j.biortech.2018.08.095
Giovana S, Padilha Elias B, TambourgiRanulfo M, Alegre, (2018) Evaluation of lipase from Burkholderia cepacia immobilized in alginate beads and application in the synthesis of banana flavor (isoamyl acetate). Chem Eng Commun 205(1):23–33. https://doi.org/10.1080/00986445.2017.1370707
Ulu A, Koytepe S, Ates B (2016) Design of starch functionalized biodegradable P(MAA-co-MMA) as carrier matrix for l-asparaginase immobilization. Carbohydr Polym 153:559–572. https://doi.org/10.1016/j.carbpol.2016.08.019
Monajati M, Borandeh S, Hesami A, Mansouri D, Tamaddon AM (2018) Immobilization of l-asparaginase on aspartic acid functionalized graphene oxide nanosheet: enzyme kinetics and stability studies. Chem Eng 354:1153–1163. https://doi.org/10.1016/j.cej.2018.08.058
Tarhan T, Ulu A, Saricam M, Culha M, Ates B (2020) Maltose functionalized magnetic core/shell Fe3O4@Au nanoparticles for an efficient l-asparaginase immobilization. Int J Biol Macromol 142:443–451. https://doi.org/10.1016/j.ijbiomac.2019.09.116
Abd El Baky HH, El Baroty GS (2020) Spirulina maxima l-asparaginase: immobilization, antiviral and antiproliferation activities. Recent Pat Biotechnol 14:2. https://doi.org/10.2174/1872208313666191114151344
Orhan H, AktaşUygun D (2020) Immobilization of l-asparaginase on magnetic nanoparticles for cancer treatment. Al Biochem Biotechnol. https://doi.org/10.1007/s12010-020-03276-z
Raybould MIJ, Marks C, Krawczyk K, Taddese B, Nowak J, Lewis AP, Bujotzek A, Shi J, Deane CM (2019) Five computational developability guidelines for therapeutic antibody profiling. Proc Natl Acad Sci USA 116(10):4025–4030. https://doi.org/10.1073/pnas.1810576116
Yu J, Shi S, Zhang F, Chen G, Cao M (2019) Predgly: predicting lysine glycation sites for homo sapiens based on XGboost feature optimization. Bioinformatics 35:2749–2756. https://doi.org/10.1093/bioinformatics/bty1043
Islam MM, Saha S, Rahman MM, Shatabda S, Farid DM, Dehzangi A (2018) IProtGly-SS: identifying protein glycation sites using sequence and structure based features. Proteins 86:777–789. https://doi.org/10.1002/prot.25511
Ju Z, Sun J, Li Y, Wang L (2017) Predicting lysine glycation sites using bi-profile bayes feature extraction. Comput Biol Chem 71:98–103. https://doi.org/10.1016/j.compbiolchem.2017.10.004
Xu Y, Li L, Ding J, Wu LY, Mai G, Zhou F (2017) Gly-PseAAC: identifying protein lysine glycation through sequences. Gene 602:1–7. https://doi.org/10.1016/j.gene.2016.11.021
Reddy HM, Sharma A, Dehzangi A, Shigemizu D, Chandra AA, Tsunoda T (2019) Glystruct: glycation prediction using structural properties of amino acid residues. BMC Bioinform 19:547. https://doi.org/10.1186/s12859-018-2547-x
Li F, Li C, Revote J, Zhang Y, Webb GI, Li J, Song J, Lithgow T (2016) GlycoMinestruct: a new bioinformatics tool for highly accurate mapping of the human N-linked and O-linked glycoproteomes by incorporating structural features. Sci Rep 6:34595. https://doi.org/10.1038/srep34595
Akmal MA, Rasool N, Khan YD (2017) Prediction of N-linked glycosylation sites using position relative features and statistical moments. PLoS ONE 12:e0181966. https://doi.org/10.1371/journal.pone.0181966
Chuang GY, Boyington JC, Joyce MG, Zhu J, Nabel GJ, Kwong PD, Georgiev I (2012) Computational prediction of N-linked glycosylation incorporating structural properties and patterns. Bioinformatics 28:2249–2255. https://doi.org/10.1093/bioinformatics/bts426
Sydow JF, Lipsmeier F, Larraillet V, Hilger M, Mautz B, Mølhøj M, Kuentzer J, Klostermann S, Schoch J, Voelger HR (2014) Structure-based prediction of asparagine and aspartate degradation sites in antibody variable regions. PLoS ONE 9:e100736. https://doi.org/10.1371/journal.pone.0100736
Aledo JC, Cantón FR, Veredas FJ (2017) A machine learning approach for predicting methionine oxidation sites. BMC Bioinform 18:430. https://doi.org/10.1186/s12859-017-1848-9
Agrawal NJ, Dykstra A, Yang J, Yue H, Nguyen X, Kolvenbach C, Angell N (2018) Prediction of the hydrogen peroxide-induced methionine oxidation propensity in monoclonal antibodies. J Pharm Sci 107:1282–1289. https://doi.org/10.1016/j.xphs.2018.01.002
Sankar K, Hoi KH, Yin Y, Ramachandran P, Andersen N, Hilderbrand A, McDonald P, Spiess C, Zhang Q (2018) Prediction of methionine oxidation risk in monoclonal antibodies using a machine learning method. MAbs 10:1281–1290. https://doi.org/10.1080/19420862.2018.1518887
Yan Q, Huang M, Lewis MJ, Hu P (2018) Structure based prediction of asparagine deamidation propensity in monoclonal antibodies. MAbs 10:901–912. https://doi.org/10.1080/19420862.2018.1478646
Lorenzo JR, Alonso LG, Sánchez IE (2015) Prediction of spontaneous protein deamidation from sequence-derived secondary structure and intrinsic disorder. PLoS ONE 10:e0145186. https://doi.org/10.1371/journal.pone.0145186
Jia L, Sun Y (2017) Protein asparagine deamidation prediction based on structures with machine learning methods. PLoS ONE 12:e0181347. https://doi.org/10.1371/journal.pone.0181347
Baskar G, Renganathan S (2010) Optimization of l-asparaginase production by Aspergillus terreus MTCC 1782 using response surface methodology and artificial neural network-linked genetic algorithm. Asia-Pac J Chem Eng 7(2):212–220. https://doi.org/10.1002/apj.520
Baskar G, Renganathan S (2011) Statistical and evolutionary optimisation of operating conditions for enhanced production of fungal l-asparaginase. Chem Pap 65(6):798–804. https://doi.org/10.2478/s11696-011-0072-8
Sushma C, Anand AP, Veeranki VD (2017) Enhanced production of glutaminase free l-asparaginase II by Bacillus subtilis WB800N through media optimization. Korean J Chem Eng 34:2901–2915. https://doi.org/10.1007/s11814-017-0211-1
Nemudryi AA, Valetdinova KR, Medvedev SP, Zakian SM (2014) TALEN and CRISPR/Cas genome editing systems: tools of discovery. Acta Nat. 6(3):19–40. https://doi.org/10.32607/20758251-2014-6-3-19-40
Chan LN, Chen Z, Braas D, Lee JW, Xiao G, Geng H, Cosgun KN, Hurtz C, Shojaee S, Cazzaniga V et al (2017) Metabolic gatekeeper function of B-lymphoid transcription factors. Nature 542(7642):479–483. https://doi.org/10.1038/nature21076
Navarro JM, Touzart A, Pradel LC, Loosveld M, Koubi M, Fenouil R, Le Noir S, Maqbool MA, Morgado E, Gregoire C et al (2015) Site- and allele-specific polycomb dysregulation in T-cell leukaemia. Nat Commun 6:6094. https://doi.org/10.1038/ncomms7094
Rahman S, Magnussen M, Leon TE, Farah N, Li Z, Abraham BJ, Alapi KZ, Mitchell RJ, Naughton T, Fielding AK et al (2017) Activation of the LMO2 oncogene through a somatically acquired neomorphic promoter in T-cell acute lymphoblastic leukemia. Blood 129(24):3221–3226. https://doi.org/10.1182/blood-2016-09-742148
Reimer J, Knoss S, Labuhn M, Charpentier EM, Gohring G, Schlegelberger B, Klusmann JH, Heckl D (2017) CRISPR-Cas9-induced t(11;19)/MLL-ENL translocations initiate leukemia in human hematopoietic progenitor cells in vivo. Haematologica 102(9):1558–1566. https://doi.org/10.3324/haematol.2017.164046
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339(6121):823–826. https://doi.org/10.1126/science.1232033
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339(6121):819–823. https://doi.org/10.1126/science.1231143
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
RKN, SE: conceptualization & supervision, SAS: writing-original draft preparation.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Suresh, S.A., Ethiraj, S. & Rajnish, K. A systematic review of recent trends in research on therapeutically significant l-asparaginase and acute lymphoblastic leukemia. Mol Biol Rep 49, 11281–11287 (2022). https://doi.org/10.1007/s11033-022-07688-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07688-4