Abstract
Background
The c-myc oncogene, which causes glutamine dependence in triple negative breast cancers (TNBC), is also the target of one of the signaling pathways affected by β-Escin.
Methods and results
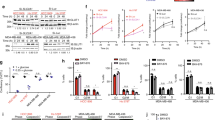
We sought to determine how c-myc protein affects glutamine metabolism and the proteins, glutamine transporter alanine-serine-cysteine 2 (ASCT2) and glutaminase (GLS1), in β-Escin-treated MDA-MB-231 cells using glutamine uptake and western blot analysis. Cell viability, colony formation, migration and apoptosis were also evaluated in MDA-MB-231 cells in response to β-Escin treatment using MTS, colony forming, wound healing, and Annexin-V assay. We determined that β-Escin decreased glutamine uptake and reduced c-myc and GLS1 protein expressions and increased the expression of ASCT2. In addition, this inhibition of glutamine metabolism decreased cell proliferation, colony formation and migration, and induced apoptosis.
Conclusions
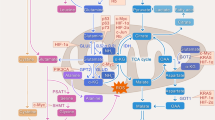
In this study, it was suggested that β-Escin inhibits glutamine metabolism via c-myc in MDA-MB-231 cells, and it is thought that as a result of interrupting the energy supply in these cells via c-myc, it results in a decrease in the carcinogenic properties of the cells. Consequently, β-Escin may be promising as a therapeutic agent for glutamine-dependent cancers.




Similar content being viewed by others
Code availability
Not applicable.
References
Cappelletti V, Iorio E, Miodini P, Silvestri M, Dugo M, Daidone MG (2017) Metabolic Footprints and Molecular Subtypes in Breast Cancer. Disease markers 7687851
El Ansari R, McIntyre A, Craze ML, Ellis IO, Rakha EA, Green AR (2018) Altered glutamine metabolism in breast cancer; subtype dependencies and alternative adaptations. Histopathology 72:183–190
Kim S, Kim DH, Jung WH, Koo JS (2013) Expression of glutamine metabolism-related proteins according to molecular subtype of breast cancer. Endocrine-related Cancer 20:339–348
Delgir S, Bastami M, Ilkhani K, Safi A, Seif F, Alivand MR (2021) The pathways related to glutamine metabolism, glutamine inhibitors and their implication for improving the efficiency of chemotherapy in triple-negative breast cancer. Mutat Res Reviews Mutat Res 787:108366
Lampa M, Arlt H, He T, Ospina B, Reeves J et al (2017) Glutaminase is essential for the growth of triple-negative breast cancer cells with a deregulated glutamine metabolism pathway and its suppression synergizes with mTOR inhibition. PLoS ONE 12:e0185092
Edwards DN, Ngwa VM, Raybuck AL, Wang S, Hwang Y et al (2021) Selective glutamine metabolism inhibition in tumor cells improves antitumor T lymphocyte activity in triple-negative breast cancer. The Journal of clinical investigation 131
Masisi BK, El Ansari R, Alfarsi L, Rakha EA, Green AR, Craze ML (2020) The role of glutaminase in cancer. Histopathology 76:498–508
Mates JM, Segura JA, Martin-Rufian M et al (2013) Glutaminase isoenzymes as key regulators in metabolic and oxidative stress against cancer. Curr Mol Med 13:514–534
Katt WP, Lukey MJ, Cerione RA (2017) A tale of two glutaminases: homologous enzymes with distinct roles in tumorigenesis. Future Med Chem 9:223–243
Martín-Rufián M, Tosina M, Campos-Sandoval JA, Manzanares E, Lobo C et al (2012) Mammalian glutaminase Gls2 gene encodes two functional alternative transcripts by a surrogate promoter usage mechanism. PLoS ONE 7:e38380
Gao P, Tchernyshyov I, Chang T-C, Lee Y-S, Kita K et al (2009) c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458:762–765
Shukla SK, Purohit V, Mehla K, Gunda V, Chaika NV et al (2017) MUC1 and HIF-1alpha signaling crosstalk induces anabolic glucose metabolism to impart gemcitabine resistance to pancreatic cancer. Cancer Cell 32:71–87 e7
Anso E, Mullen AR, Felsher DW, Matés JM, DeBerardinis RJ, Chandel NS (2013) Metabolic changes in cancer cells upon suppression of MYC. Cancer & metabolism 1:7
Yuneva MO, Fan TW, Allen TD, Higashi RM, Ferraris DV et al (2012) The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metabol 15:157–170
Martinez-Outschoorn UE, Peiris-Pages M, Pestell RG, Sotgia F, Lisanti MP (2017) Cancer metabolism: a therapeutic perspective. Nat Rev Clin Oncol 14:11–31
Ezberci F, Unal E (2018) Aesculus Hippocastanum (Aescin, Horse Chestnut) in the Management of Hemorrhoidal Disease. Turk J Colorectal Dis 28:54–57
Raafat M, Kamel AA, Shehata AH, Ahmed AF, Bayoumi AMA et al (2022) Aescin Protects against Experimental Benign Prostatic Hyperplasia and Preserves Prostate Histomorphology in Rats via Suppression of Inflammatory Cytokines and COX-2. Pharmaceuticals 15
Pittler MH, Ernst E (2012) Horse chestnut seed extract for chronic venous insufficiency. Cochrane Database Syst Rev 11:CD003230
Gallelli L (2019) Escin: a review of its anti-edematous, anti-inflammatory, and venotonic properties. Drug Des Devel Ther 13:3425–3437
Yuan S-Y, Cheng C-L, Wang S-S, Ho H-C, Chiu K-Y et al (2017) Escin induces apoptosis in human renal cancer cells through G2/M arrest and reactive oxygen species-modulated mitochondrial pathways. Oncol Rep 37:1002–1010
Mojžišová G, Kello M, Pilátová M, Tomečková V, Vašková J et al (2016) Antiproliferative effect of β-escin-an in vitro study. Acta Biochim Pol 63:79–87
Zhang Z, Gao J, Cai X, Zhao Y, Wang Y et al (2011) Escin sodium induces apoptosis of human acute leukemia Jurkat T cells. Phytother Res 25:1747–1755
Niu Y-P, Li L-D, Wu L-M (2008) Beta-aescin: a potent natural inhibitor of proliferation and inducer of apoptosis in human chronic myeloid leukemia K562 cells in vitro. Leuk Lymphoma 49:1384–1391
Çiftçi GA, Işcan A, Kutlu M (2015) Escin reduces cell proliferation and induces apoptosis on glioma and lung adenocarcinoma cell lines. Cytotechnology 67:893–904
Wang Z, Chen Q, Li B, Xie JM, Yang XD et al (2018) Escin-induced DNA damage promotes escin-induced apoptosis in human colorectal cancer cells via p62 regulation of the ATM/gammaH2AX pathway. Acta Pharmacol Sin 39:1645–1660
Zhu J, Yu W, Liu B, Wang Y, Shao J et al (2017) Escin induces caspase-dependent apoptosis and autophagy through the ROS/p38 MAPK signalling pathway in human osteosarcoma cells in vitro and in vivo. Cell Death Dis 8:e3113
Wang YW, Wang SJ, Zhou YN, Pan SH, Sun B (2012) Escin augments the efficacy of gemcitabine through down-regulation of nuclear factor-kappaB and nuclear factor-kappaB-regulated gene products in pancreatic cancer both in vitro and in vivo. J Cancer Res Clin Oncol 138:785–797
Tan SM, Li F, Rajendran P, Kumar AP, Hui KM, Sethi G (2010) Identification of beta-escin as a novel inhibitor of signal transducer and activator of transcription 3/Janus-activated kinase 2 signaling pathway that suppresses proliferation and induces apoptosis in human hepatocellular carcinoma cells. J Pharmacol Exp Ther 334:285–293
Dang CV, Le A, Gao P (2009) MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin cancer research: official J Am Association Cancer Res 15:6479–6483
Liu W, Le A, Hancock C, Lane AN, Dang CV et al (2012) Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci USA 109:8983–8988
Hamurcu Z, Delibasi N, Gecene S, Sener EF, Donmez-Altuntas H et al (2018) Targeting LC3 and Beclin-1 autophagy genes suppresses proliferation, survival, migration and invasion by inhibition of Cyclin-D1 and uPAR/Integrin beta1/ Src signaling in triple negative breast cancer cells. J Cancer Res Clin Oncol 144:415–430
Hamurcu Z, Delibasi N, Nalbantoglu U, Sener EF, Nurdinov N et al (2019) FOXM1 plays a role in autophagy by transcriptionally regulating Beclin-1 and LC3 genes in human triple-negative breast cancer cells. J Mol Med 97:491–508
Unal TD, Hamurcu Z, Delibasi N, Cinar V, Guler A et al (2021) Thymoquinone Inhibits Proliferation and Migration of MDA-MB-231 Triple Negative Breast Cancer Cells by Suppressing Autophagy, Beclin-1 and LC3. Anticancer Agents Med Chem 21:355–364
Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB (2008) Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev 18:54–61
Chang CF, Diers AR, Hogg N (2015) Cancer cell metabolism and the modulating effects of nitric oxide. Free Radic Biol Med 79:324–336
Timmerman LA, Holton T, Yuneva M, Louie RJ, Padro M et al (2013) Glutamine sensitivity analysis identifies the xCT antiporter as a common triple-negative breast tumor therapeutic target. Cancer Cell 24:450–465
Wang Y, Xu X, Zhao P, Tong B, Wei Z, Dai Y (2016) Escin Ia suppresses the metastasis of triple-negative breast cancer by inhibiting epithelial-mesenchymal transition via down-regulating LOXL2 expression. Oncotarget 7:23684–23699
Kwak H, An H, Alam MB, Choi WS, Lee SY, Lee SH (2018) Inhibition of Migration and Invasion in Melanoma Cells by beta-Escin via the ERK/NF-kappaB Signaling Pathway. Biol Pharm Bull 41:1606–1610
Kenny HA, Hart PC, Kordylewicz K, Lal M, Shen M et al (2021) The Natural Product beta-Escin Targets Cancer and Stromal Cells of the Tumor Microenvironment to Inhibit Ovarian Cancer Metastasis. Cancers 13
Omi K, Matsuo Y, Ueda G, Aoyama Y, Kato T et al (2021) Escin inhibits angiogenesis by suppressing interleukin8 and vascular endothelial growth factor production by blocking nuclear factor kappaB activation in pancreatic cancer cell lines. Oncology reports 45
Paneerselvam C, Ganapasam S (2020) beta-Escin alleviates cobalt chloride-induced hypoxia-mediated apoptotic resistance and invasion via ROS-dependent HIF-1alpha/TGF-beta/MMPs in A549 cells. Toxicol Res 9:191–201
Huang GL, Shen DY, Cai CF, Zhang QY, Ren HY, Chen QX (2015) beta-escin reverses multidrug resistance through inhibition of the GSK3beta/beta-catenin pathway in cholangiocarcinoma. World J Gastroenterol 21:1148–1157
van Geldermalsen M, Wang Q, Nagarajah R, Marshall AD, Thoeng A et al (2016) ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene 35:3201–3208
Acknowledgements
This work was financially supported by the Scientific and Technological Research Council of Turkey (TUBITAK). Research Grant (Grant number: 119S227).
Author information
Authors and Affiliations
Contributions
All authors contributed equally to this work.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest or other disclosures to report.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
AKAR, S., DONMEZ-ALTUNTAS, H. & HAMURCU, Z. β-Escin reduces cancer progression in aggressive MDA-MB-231 cells by inhibiting glutamine metabolism through downregulation of c-myc oncogene. Mol Biol Rep 49, 7409–7415 (2022). https://doi.org/10.1007/s11033-022-07536-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07536-5