Abstract
Background
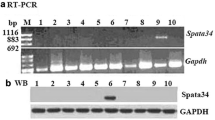
Fascins belong to a family of actin-bundling proteins that are involved in a wide range of biological functions. FSCN3, a newly identified testis-specific actin-bundling protein, is specifically expressed in elongated spermatids. However, its in vivo function in mouse spermiogenesis remains unknown.
Methods and results
We generated Fscn3 knockout mice through CRISPR/Cas9 gene-editing technology. Fscn3−/− mice displayed normal testis morphology and testis to bodyweight ratio, and sperm concentrations did not differ significantly between Fscn3+/+ and Fscn3−/− mice. Fertility assays consistently revealed that Fscn3−/− mice are completely fertile and their reproductive status does not differ from that of wild-type. Moreover, hematoxylin and eosin staining of the testis sections of Fscn3−/− mice detected various germ cells, ranging from spermatogonia to mature spermatozoa. Furthermore, the swimming velocity of the sperm of Fscn3−/− mice was comparable to that of their wild-type littermates. Both Fscn3+/+ and Fscn3−/−mice had normal sperm morphology, indicating that the disruption of Fscn3 does not affect sperm morphology. The analysis of meiotic prophase I progression demonstrated normal prophase-I phases (leptonema to diplonema) in both Fscn3+/+ and Fscn3−/− mice, suggesting that Fscn3 is not essential for meiosis I.
Conclusion
Our study provides the first evidence that FSCN3 is a testis-specific actin-bundling protein that is not required for mouse spermatogenesis. Our results will help reproductive biologists focus their efforts on genes that are crucial for fertility and avoid research duplication.




Similar content being viewed by others
References
Pashaei M et al (2020) The second mutation of SYCE1 gene associated with autosomal recessive nonobstructive azoospermia. J Assist Reprod Genet 37:451–458
Schilit SLP et al (2020) SYCP2 translocation-mediated dysregulation and frameshift variants cause human male infertility. Am J Hum Genet 106:41–57
Bolor H et al (2009) Mutations of the SYCP3 gene in women with recurrent pregnancy loss. Am J Hum Genet 84:14–20
Nakamura L (2013) Cybertypes: race, ethnicity, and identity on the Internet. Routledge, London
Chocu S, Calvel P, Rolland AD, Pineau C (2012) Spermatogenesis in mammals: proteomic insights. Syst Biol Reprod Med 58:179–190
Wang H, Yuan Q, Niu M, Zhang W, Wen L, Fu H, He Z (2018) Transcriptional regulation of P63 on the apoptosis of male germ cells and three stages of spermatogenesis in mice. Cell 9(2):1–16
Schultz N, Hamra FK, Garbers DL (2003) A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc Natl Acad Sci USA 100(21):12201–12206
Legrand JMD, Chan A-L, La HM, Rossello FJ, Änkö M-L, FullerPace FV, Hobbs RM (2019) DDX5 plays essential transcriptional and post-transcriptional roles in the maintenance and function of spermatogonia. Nat Commun 10:2278
Miyata H, Satouh Y, Mashiko D, Muto M, Nozawa K, Shiba K, Fujihara Y, Isotani A, Inaba K, Ikawa M (2015) Sperm calcineurin inhibition prevents mouse fertility with implications for male contraceptive. Science 350:442–445
Young S, Miyata H, Satouh Y, Kato H, Nozawa K, Isotani A, Aitken RJ, Baker MA, Ikawa M (2015) CRISPR/Cas9-mediated rapid generation of multiple mouse lines identified Ccdc63 as essential for spermiogenesis. Int J Mol Sci 16:24732–24750
Lu Y, Oura S, Matsumura T, Oji A, Sakurai N et al (2019) CRISPR/Cas9-mediated genome editing reveals 30 testis-enriched genes dispensable for male fertility in mice. Biol Reprod 101:501–511
Park S, Shimada K, Fujihara Y, Xu Z, Shimada K et al (2020) CRISPR/Cas9-mediated genome-edited mice reveal 10 testis-enriched genes are dispensable for male fecundity. Biol Reprod 103:195–204
Sun J, Lu Y, Nozawa K, Xu Z, Morohoshi A et al (2020) CRISPR/Cas9-based genome editing in mice uncovers 13 testis- or epididymis-enriched genes individually dispensable for male reproduction. Biol Reprod 103:183–194
Miyata H, Castaneda JM, Fujihara Y, Yu Z, Archambeault DR et al (2016) Genome engineering uncovers 54 evolutionarily conserved and testis-enriched genes that are not required for male fertility in mice. Proc Natl Acad Sci USA 113:7704–7710
Young SA, Miyata H, Satouh Y, Aitken RJ, Baker MA, Ikawa M (2016) CABYR is essential for fibrous sheath integrity and progressive motility in mouse spermatozoa. J Cell Sci 129:4379–4387
Xia X, Zhou X, Quan Y, Hu Y, Xing F, Li Z, Xu B, Xu C, Zhang A (2019) Germline deletion of Cdyl causes teratozoospermia and progressive infertility in male mice. Cell Death Dis 10(3):229
Khan M, Jabeen N, Khan T, Hussain HMJ, Ali A, Khan R, Jiang L, Li T, Tao Q, Zhang X, Yin H, Yu C, Jiang X, Shi Q (2018) The evolutionarily conserved genes: Tex37, Ccdc73, Prss55 and Nxt2 are dispensable for fertility in mice. Sci Rep 8(1):4975
Jiang XH, Bukhari I, Zheng W, Yin S, Wang Z, Cooke HJ, Shi QH (2014) Blood-testis barrier and spermatogenesis: lessons from genetically-modified mice. Asian J Androl 16:572
Abbasi F, Miyata H, Ikawa M (2017) Revolutionizing male fertility factor research in mice by using the genome editing tool CRISPR/Cas9. Reprod Med Biol 17(1):3–10
Bryan J, Edwards R, Matsudaira P, Otto J, Wulfkuhle J (1993) Fascin, an echinoid actin-bundling protein, is a homolog of the Drosophila singed gene product. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.90.19.9115
Edwards RA, Bryan J (1995) Fascins, a family of actin bundling proteins. Cell Motil Cytoskelet 32:1–9
Yamashiro-Matsumura S, Matsumura F (1985) Purification and characterization of an F-actin-bundling 55-kilodalton protein from HeLa cells. J Biol Chem 260(8):5087–5097
Tubb B, Mulholland DJ, Vogl W, Lan ZJ, Niederberger C, Cooney A et al (2002) Testis fascin (FSCN3): a novel paralog of the actin-bundling protein fascin expressed specifically in the elongate spermatid head. Exp Cell Res 275:92–109
Kollers S, Day A, Rocha D (2006) Characterization of the porcine FSCN3 gene: cDNA cloning, genomic structure, mapping and polymorphisms. Cytogenet Genome Res 115:189–192
Malcher A, Rozwadowska N, Stokowy T, Kolanowski T, Jedrzejczak P, Zietkowiak W, Kurpisz M (2013) Potential biomarkers of nonobstructive azoospermia identified in microarray gene expression analysis. Fertil Steril. https://doi.org/10.1016/j.fertnstert.2013.07.1999
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, New York
Jiang L, Li T, Zhang X, Zhang B, Yu C, Li Y et al (2017) RPL10L is required for male meiotic division by compensating for RPL10 during meiotic sex chromosome inactivation in mice. Curr Biol 27(10):1498–505
Jiang X, Ma T, Zhang Y, Zhang H, Yin S, Zheng W et al (2015) Specific deletion of Cdh2 in Sertoli cells leads to altered meiotic progression and subfertility of mice. Biol Reprod. https://doi.org/10.1095/biolreprod.114.126334
Smith J, Grizot S, Arnould S, Duclert A, Epinat JC, Chames P, Prieto J, Redondo P, Blanco FJ, Bravo J, Montoya G, Pâques F, Duchateau P (2006) A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res 34(22):e149
Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153(4):910–918. https://doi.org/10.1016/j.cell.2013.04.025
Liu F, Jin S, Li N, Liu X, Wang H, Li J (2011) Comparative and functional analysis of testisspecific genes. Biol Pharm Bull 34(1):28–35
Ahmed EA, Sfeir A, Takai H, Scherthan H (2013) Ku70 and non-homologous end joining protect testicular cells from DNA damage. J Cell Sci 126(Pt 14):3095–3104
Hu X, Shen B, Liao S, Ning Y, Ma L, Chen J et al (2017) Gene knockout of Zmym3 in mice arrests spermatogenesis at meiotic metaphase with defects in spindle assembly checkpoint. Cell Death Dis 8(6):e2910
Acknowledgements
This work was supported by the National Key Research and Developmental Program of China (Grant Nos. 2018YFC1003403 and 2016YFC1000600), the National Natural Science Foundation of China (Grant Nos. 31890780 and 31630050), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB19000000), and the Fundamental Research Funds for the Central Universities (Grant No. YD2070002006).
Funding
This work was supported by National Basic Research Program of China (973 Program) [Grant Nos. 2018YFC1004700 and 2016YFC1000600], Innovative Research Group Project of the National Natural Science Foundation of China [Grant Nos. 31890780, 31630050], the Strategic Priority Research Program of the Chinese Academy of Sciences [Grant No. XDB19000000], Fundamental Research Funds for the Central Universities [Grant No. YD2070002006].
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: QS, HA. Performed the experiments: HA, SD, AUU, KK. Analyzed the data: HA, SD, AUU, IA. Wrote the paper: HA, SD, AUU.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
All experiments and examination on laboratory animals were conducted according to the institutional rules of the Institutional Animal Care Committee of the University of Science and Technology of China.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ali, H., Unar, A., Dil, S. et al. Testis-specific fascin component FSCN3 is dispensable for mouse spermatogenesis and fertility. Mol Biol Rep 49, 6261–6268 (2022). https://doi.org/10.1007/s11033-022-07429-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07429-7