Abstract
TP53 functions primarily as a tumor suppressor, controlling a myriad of signalling pathways that prevent a cell from undergoing malignant transformation. This tumor suppressive function requires an activation and stabilization of TP53 in response to cell stressors. However, besides its cancer-preventive functions, TP53 is also known to be involved in diverse cellular processes including metabolism, reproduction, stem cell renewal and development. Indeed, several lines of evidence strongly suggest that TP53 plays crucial role in diabetes. A number of studies have evaluated the association of genetic alterations (single nucleotide variations) in TP53 gene with the development of diabetes. However, the results have not been consistent. The aim of this study was to evaluate whether the C/G polymorphism at codon 72 (Pro72/Arg72), located in exon 4 of TP53, is associated with type 2 diabetes in South Indian population. A total of 74 type 2 diabetic patients and 54 non-diabetic subjects were screened. None of the three genotypes, namely C/C (Pro/Pro), C/G (Pro/Arg), and G/G (Arg/Arg) was found to be significantly associated with type 2 diabetes in our study group. The findings of this study indicate that TP53 codon 72 polymorphism is not associated with increased risk of type 2 diabetes in South Indian population. Further studies with a large cohort size would be necessary to corroborate the observations of this study. Nevertheless, this study represents the first genetic analysis of TP53 variants in South Indian type 2 diabetic patients.

Similar content being viewed by others
Abbreviations
- SNP:
-
Single nucleotide polymorphism
- RFLP:
-
Restriction fragment length polymorphism
- BMI:
-
Body mass index
- DM:
-
Diabetes mellitus
References
Chakraborty A, Uechi T, Kenmochi N (2011) Guarding the ‘translation apparatus’: defective ribosome biogenesis and the p53 signaling pathway. WIREs RNA 2(4):507–522
Kung CP, Murphy ME (2016) The role of the p53 tumor suppressor in metabolism and diabetes. J Endocrinol 231(2):R61
Brownlee M (2005) The pathobiology of diabetic complications: a unifying mechanism. Diabetes 54(6):1615–1625
Labuschagne CF, Zani F, Vousden KH (2018) Control of metabolism by p53 cancer and beyond. Biochim Biophys Acta Rev Cancer 1870(1):32–42
Lacroix M, Riscal R, Arena G, Linares LK, Le Cam L (2020) Metabolic functions of the tumor suppressor p53: implications in normal physiology, metabolic disorders, and cancer. Mol Met 33:2–22
Wang X, Zhao X, Gao X, Mei Y, Wu M (2013) A new role of p53 in regulating lipid metabolism. J Mol Cell Biol 5(2):147–150
Secchiero P, Toffoli B, Melloni E, Agnoletto C, Monasta L, Zauli G (2013) The MDM2 inhibitor Nutlin-3 attenuates streptozotocin-induced diabetes mellitus and increases serum level of IL-12p40. Acta diabetol 50(6):899–906
Armata HL, Golebiowski D, Jung DY, Ko HJ, Kim JK, Sluss HK (2010) Requirement of the ATM/p53 tumor suppressor pathway for glucose homeostasis. Mol Cell Biol 30(24):5787–5794
Franck D, Tracy L, Armata HL, Delaney CL, Jung DY, Ko HJ, Ong H, Kim JK, Scrable H, Sluss HK (2013) Glucose tolerance in mice is linked to the dose of the p53 transactivation domain. Endocr Res 38(3):139–150
Lin HY, Huang CH, Wu WJ, Chang LC, Lung FW (2008) TP53 codon 72 gene polymorphism paradox in associated with various carcinoma incidences, invasiveness and chemotherapy responses. Int J Biol Sci 4(4):248
Bonfigli AR, Sirolla C, Testa R, Cucchi M, Spazzafumo L, Salvioli S, Ceriello A, Olivieri F, Festa R, Procopio AD, Brandoni G (2013) The p53 codon 72 (Arg72Pro) polymorphism is associated with the degree of insulin resistance in type 2 diabetic subjects: a cross-sectional study. Acta diabetol 50(3):429–436
Bitti MM, Saccucci P, Bottini E, Gloria-Bottini F (2010) p53 codon 72 polymorphism and type 1 diabetes mellitus. J Pediatr Endocrinol Metab 23(3):291–292
Gloria-Bottini F, Banci M, Saccucci P, Magrini A, Bottini E (2011) Is there a role of p53 codon 72 polymorphism in the susceptibility to type 2 diabetes in overweight subjects? A study in patients with cardiovascular diseases. Diabetes Res Clin Pract 91(3):64–67
Neri A, Gloria-Bottini F, Banci M, Magrini A, Bottini E (2017) Cigarette smoking increases the effect of *Arg/*Arg genotype of P53 codon 72 on the susceptibility to type 2 diabetes in overweight subjects. J Tob Stimul Dis 1(1):013–015
Gaulton KJ, Willer CJ, Li Y, Scott LJ, Conneely KN, Jackson AU, Duren WL, Chines PS, Narisu N, Bonnycastle LL, Luo J (2008) Comprehensive association study of type 2 diabetes and related quantitative traits with 222 candidate genes. Diabetes 57(11):3136–3144
Leroy B, Anderson M, Soussi T (2014) TP53 mutations in human cancer: database reassessment and prospects for the next decade. Hum Mutat 35(6):672–688
Dumont P, Leu JJ, Della Pietra AC, George DL, Murphy M (2003) The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet 33(3):357–365
Kung CP, Khaku S, Jennis M, Zhou Y, Murphy ME (2015) Identification of TRIML2, a novel p53 target, that enhances p53 SUMOylation and regulates the transactivation of proapoptotic genes. Mol Cancer Res 13(2):250–262
Spitsina EV, NIu I, Chudakova DA, Nikitin AG, Svetlova GN, Soluianova TN, Strokov IA, Nosikov VV (2007) Association of polymorphous markers Pro72Arg and C (−594) CC OF TP53 gene with diabetic polyneuropathy in patients with type 1 diabetes mellitus living in Moscow. Mol Biol 41(6):989–993
Bitti ML, Saccucci P, Capasso F, Piccinini S, Angelini F, Rapini N, Porcari M, Arcano S, Petrelli A, Del Duca E, Bottini E (2011) Genotypes of p53 codon 72 correlate with age at onset of type 1 diabetes in a sex-specific manner. J Pediatr Endocrinol Metab 24(7–8):437–439
Burgdorf KS, Grarup N, Justesen JM, Harder MN, Witte DR, Jørgensen T, Sandbæk A, Lauritzen T, Madsbad S, Hansen T, Pedersen O (2011) Studies of the association of Arg72Pro of tumor suppressor protein p53 with type 2 diabetes in a combined analysis of 55,521 Europeans. PLoS One 6(1):e15813
Sliwinska A, Kasznicki J, Kosmalski M, Mikołajczyk M, Rogalska A, Przybylowska K, Majsterek I, Drzewoski J (2017) Tumour protein p53 is linked with type 2 diabetes mellitus. Indian J Med Res 146(2):237–243
Słomiński B, Skrzypkowsk M, Ryba- Stanisławowska M, Myśliwiec M, Trzonkowski P (2021) Associations of TP53 codon 72 polymorphism with complications and comorbidities in patients with type 1diabetes. J Mol Med 99:675–683
Acknowledgements
The authors are grateful to the patients and the healthy controls who have agreed to take part in this study. The authors thank Nitte (Deemed to be University) for providing resource and infrastructural support. The authors also acknowledge the help of Dr. Neevan D’Souza from KSHEMA in the statistical analysis of the data.
Author information
Authors and Affiliations
Contributions
HKP collected the samples and curated the clinical data. DPN performed the molecular studies and prepared the original draft. BSPD, NB and KK partly performed the experiments and analysed the data. GC participated in discussion and edited the first draft of the manuscript. SMR and HKP conceptualised the study and helped in clinical data compilation. AC supervised the work, analysed the data and edited the draft manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no conflict of interest.
Informed consent
Informed consent from the participants were obtained and the study was approved by Ethics Committee of Nitte (Deemed to be University, RefNU/CEC/2018/0).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11033_2021_6505_MOESM1_ESM.tif
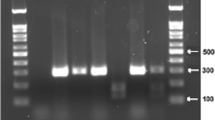
Supplementary file1 Fig. 1 An image showing agarose gel electrophoresis of PCR products of TP53 codon 72. M: 100bp DNA ladder, Lane 1-10: Amplified gene fragment of TP53 codon 72 (TIF 4256 kb)
11033_2021_6505_MOESM2_ESM.tif
Supplementary file2 Fig. 2 A representative image of RFLP analysis at codon 72 of TP53.The RFLP pattern obtained from each sample was compared with the control RFLP pattern to examine the genotype. M-100 base pair ladder; U-undigested PCR product; D-digested PCR product. Sample no. 1 and 5 showed-G/G (wildtype), 2 and 3 - G/C (heterozygous), and 4 - C/C(homozygous) (TIF 4408 kb)
Rights and permissions
About this article
Cite this article
Punja, H.K., Nanjappa, D.P., Babu, N. et al. TP53 codon 72 polymorphism and type 2 diabetes: a case–control study in South Indian population. Mol Biol Rep 48, 5093–5097 (2021). https://doi.org/10.1007/s11033-021-06505-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06505-8