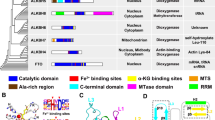
Abstract
AlkBH1 is a member of the AlkB superfamily which are kinds of Fe (II) and α-ketoglutarate (α-KG)-dependent dioxygenases. At present, only demethyltransferases FTO and AlkBH5 have relatively clear substrate studies among these members, the types and mechanisms of substrates catalysis of other members are not clear, especially the demethyltransferase AlkBH1. AlkBH1, as a demethylase, has important functions of reversing DNA methylation and repairing DNA damage. And it has become a promising target for the treatment of many cancers, the regulation of neurological and genetic related diseases. Many scholars have made important discoveries in the diversity of AlkBH1 substrates, but there is no comprehensive summary, which affects the design inhibitor target of AlkBH1. Herein, We are absorbed in the latest progress in the study of AlkBH1 substrate diversity and its relationship with human diseases. Besides, we also discuss future research directions and suggest other studies to reveal the specific catalytic effect of AlkBH1 on cancer substrates.




Similar content being viewed by others
References:
Mishina Y, He C (2006) Oxidative dealkylation DNA repair mediated by the mononuclear non-heme iron AlkB proteins. J Inorg Biochem 100:670–678
Fedeles BI, Singh V, Delaney JC, Li D, Essigmann JM (2015) The AlkB family of Fe(II)/alpha-ketoglutarate-dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J Biol Chem 290:20734–20742
Guengerich FP (2015) Introduction: metals in biology: alpha-ketoglutarate/Iron-dependent dioxygenases. J Biol Chem 290:20700–20701
Welford RW, Kirkpatrick JM, McNeill LA, Puri M, Oldham NJ, Schofield CJ (2005) Incorporation of oxygen into the succinate co-product of iron(II) and 2-oxoglutarate dependent oxygenases from bacteria, plants and humans. FEBS Lett 579:5170–5174
Rudkjobing LA, Eiberg H, Mikkelsen HB, Binderup ML, Bisgaard ML (2015) The analysis of a large Danish family supports the presence of a susceptibility locus for adenoma and colorectal cancer on chromosome 11q24. Fam Cancer 14:393–400
Shivange G, Kodipelli N, Anindya R (2014) A nonradioactive restriction enzyme-mediated assay to detect DNA repair by Fe(II)/2-oxoglutarate-dependent dioxygenase. Anal Biochem 465:35–37
Brickner JR, Soll JM, Lombardi PM et al (2017) A ubiquitin-dependent signalling axis specific for ALKBH-mediated DNA dealkylation repair. Nature 551:389–393
Li Q, Huang Y, Liu X, Gan J, Chen H, Yang CG (2016) Rhein inhibits AlkB repair enzymes and sensitizes cells to methylated DNA damage. J Biol Chem 291:11083–11093
Martin Carli JF, LeDuc CA, Zhang Y, Stratigopoulos G, Leibel RL (2018) FTO mediates cell-autonomous effects on adipogenesis and adipocyte lipid content by regulating gene expression via 6mA DNA modifications. J Lipid Res 59:1446–1460
Zhang X, Wei LH, Wang Y et al (2019) Structural insights into FTO’s catalytic mechanism for the demethylation of multiple RNA substrates. Proc Natl Acad Sci 116:2919–2924
Zhu Y, Zhou G, Yu X et al (2017) LC-MS–MS quantitative analysis reveals the association between FTO and DNA methylation. PLoS ONE 12:e0175849
Jin D, Guo J, Wu Y et al (2020) m(6)A demethylase ALKBH5 inhibits tumor growth and metastasis by reducing YTHDFs-mediated YAP expression and inhibiting miR-107/LATS2-mediated YAP activity in NSCLC. Mol Cancer 19:40
Chen S, Zhou L, Wang Y (2020) ALKBH5-mediated m(6)A demethylation of lncRNA PVT1 plays an oncogenic role in osteosarcoma. Cancer Cell Int 20:34
Woo HH, Chambers SK (2019) Human ALKBH3-induced m(1)A demethylation increases the CSF-1 mRNA stability in breast and ovarian cancer cells. Biochim Biophys Acta Gene Regul Mech 1862:35–46
Chen Z, Qi M, Shen B et al (2019) Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res 47:2533–2545
Yi C, Chen B, Qi B et al (2012) Duplex interrogation by a direct DNA repair protein in search of base damage. Nat Struct Mol Biol 19:671–676
Ji N, Wang X, Yin C, Peng W, Liang R (2019) CrgA protein represses AlkB2 monooxygenase and regulates the degradation of medium-to-long-chain n-alkanes in pseudomonas aeruginosa SJTD-1. Front Microbiol 10:400
Nilsen A, Fusser M, Greggains G, Fedorcsak P, Klungland A (2014) ALKBH4 depletion in mice leads to spermatogenic defects. PLoS ONE 9:e105113
Ougland R, Rognes T, Klungland A, Larsen E (2015) Non-homologous functions of the AlkB homologs. J Mol Cell Biol 7:494–504
Pilzys T, Marcinkowski M, Kukwa W et al (2019) ALKBH overexpression in head and neck cancer: potential target for novel anticancer therapy. Sci Rep 9:13249
Li Y, Zheng D, Wang F, Xu Y, Yu H, Zhang H (2019) Expression of demethylase genes, FTO and ALKBH1, is associated with prognosis of gastric cancer. Dig Dis Sci 64:1503–1513
Kawarada L, Suzuki T, Ohira T, Hirata S, Miyauchi K, Suzuki T (2017) ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res 45:7401–7415
Ma CJ, Ding JH, Ye TT, Yuan BF, Feng YQ (2019) AlkB homologue 1 demethylates N(3)-methylcytidine in mRNA of mammals. ACS Chem Biol 14:1418–1425
Xu L, Liu X, Sheng N et al (2017) Three distinct 3-methylcytidine (m(3)C) methyltransferases modify tRNA and mRNA in mice and humans. J Biol Chem 292:14695–14703
Zhang LH, Zhang XY, Hu T et al (2020) The SUMOylated METTL8 induces R-loop and tumorigenesis via m3C. iScience 23:100968
Wagner A, Hofmeister O, Rolland SG et al (2019) Mitochondrial Alkbh1 localizes to mtRNA granules and its knockdown induces the mitochondrial UPR in humans and C. elegans. J Cell Sci. https://doi.org/10.1242/jcs.223891
Haag S, Sloan KE, Ranjan N et al (2016) NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J 35:2104–2119
Auxilien S, Guerineau V, Szweykowska-Kulinska Z, Golinelli-Pimpaneau B (2012) The human tRNA m (5) C methyltransferase Misu is multisite-specific. RNA Biol 9:1331–1338
Van Haute L, Dietmann S, Kremer L et al (2016) Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat Commun 7:12039
Li Q, Li X, Tang H et al (2017) NSUN2-mediated m5C methylation and METTL3/METTL14-mediated m6A methylation cooperatively enhance p21 translation. J Cell Biochem 118:2587–2598
Dong Z, Cui H (2020) The emerging roles of RNA modifications in glioblastoma. Cancers 12:736
Sibbritt T, Patel HR, Preiss T (2013) Mapping and significance of the mRNA methylome. Wiley Interdiscip Rev RNA 4:397–422
Zhao BS, Roundtree IA, He C (2017) Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 18:31–42
Yang Y, Wang L, Han X et al (2019) RNA 5-methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA decay. Mol Cell 75:1188-1202 e1111
Chen X, Li A, Sun BF et al (2019) 5-Methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol 21:978–990
Yang X, Yang Y, Sun BF et al (2017) 5-Methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res 27:606–625
Liu F, Clark W, Luo G et al (2016) ALKBH1-mediated tRNA demethylation regulates translation. Cell 167:816-828 e816
Oerum S, Degut C, Barraud P, Tisne C (2017) m1A Post-transcriptional modification in tRNAs. Biomolecules 7:20
Li X, Xiong X, Wang K et al (2016) Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat Chem Biol 12:311–316
Grozhik AV, Olarerin-George AO, Sindelar M, Li X, Gross SS, Jaffrey SR (2019) Antibody cross-reactivity accounts for widespread appearance of m(1)A in 5′UTRs. Nat Commun 10:5126
Xiong X, Li X, Yi C (2018) N(1)-methyladenosine methylome in messenger RNA and non-coding RNA. Curr Opin Chem Biol 45:179–186
Safra M, Sas-Chen A, Nir R et al (2017) The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551:251–255
Peer E, Rechavi G, Dominissini D (2017) Epitranscriptomics: regulation of mRNA metabolism through modifications. Curr Opin Chem Biol 41:93–98
Rashad S, Han X, Sato K et al (2020) The stress specific impact of ALKBH1 on tRNA cleavage and tiRNA generation. RNA Biol 17:1092–1103
Wu L, Pei Y, Zhu Y et al (2019) Association of N(6)-methyladenine DNA with plaque progression in atherosclerosis via myocardial infarction-associated transcripts. Cell Death Dis 10:909
Lin S, Choe J, Du P, Triboulet R, Gregory RI (2016) The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell 62:335–345
Chen M, Wei L, Law CT et al (2018) RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 67:2254–2270
Dai D, Wang H, Zhu L, Jin H, Wang X (2018) N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis 9:124
Yu J, Chen M, Huang H et al (2018) Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res 46:1412–1423
Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR (2015) Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods 12:767–772
Erson-Bensan AE, Begik O (2017) m6A modification and implications for microRNAs. Microrna 6:97–101
Liu ZX, Li LM, Sun HL, Liu SM (2018) Link between m6A modification and cancers. Front Bioeng Biotechnol 6:89
Chen X, Yu C, Guo M et al (2019) Down-regulation of m6A mRNA methylation is involved in dopaminergic neuronal death. ACS Chem Neurosci 10:2355–2363
Wang P, Doxtader KA, Nam Y (2016) Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol Cell 63:306–317
Bedi RK, Huang D, Eberle SA, Wiedmer L, Sledz P, Caflisch A (2020) Small-molecule inhibitors of METTL3, the major human epitranscriptomic writer. ChemMedChem 15:744–748
Sun T, Wu R, Ming L (2019) The role of m6A RNA methylation in cancer. Biomed Pharmacother 112:108613
Tang C, Klukovich R, Peng H et al (2018) ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells. Proc Natl Acad Sci 115:E325–E333
Pilzys T, Marcinkowski M, Kukwa W, Garbicz D, Dylewska M, Ferenc K (2019) ALKBH overexpression in head and neck cancer: potential target for novel anticancer therapy. Sci Rep 9:13249
Xiao CL, Zhu S, He M et al (2018) N(6)-methyladenine DNA modification in the human genome. Mol Cell 71:306-318.e307
Fernandes GFS, Silva GDB, Pavan AR, Chiba DE, Chin CM, Dos Santos JL (2017) Epigenetic regulatory mechanisms induced by resveratrol. Nutrients 9:1201
Luo GZ, Blanco MA, Greer EL, He C, Shi Y (2015) DNA N(6)-methyladenine: a new epigenetic mark in eukaryotes? Nat Rev Mol Cell Biol 16:705–710
Zhang G, Huang H, Liu D et al (2015) N6-methyladenine DNA modification in drosophila. Cell 161:893–906
Zhou C, Liu Y (2016) DNA N(6)-methyladenine demethylase ALKBH1 enhances osteogenic differentiation of human MSCs. Bone Res 4:16033
Xie Q, Wu TP, Gimple RC et al (2018) N(6)-methyladenine DNA modification in glioblastoma. Cell 175:1228-1243.e1220
Zhang M, Yang S, Nelakanti R et al (2020) Mammalian ALKBH1 serves as an N(6)-mA demethylase of unpairing DNA. Cell Res 30:197–210
Tian LF, Liu YP, Chen L et al (2020) Structural basis of nucleic acid recognition and 6mA demethylation by human ALKBH1. Cell Res 30:272–275
Mielecki D, Zugaj DL, Muszewska A et al (2012) Novel AlkB dioxygenases–alternative models for in silico and in vivo studies. PLoS ONE 7:e30588
Wu TP, Wang T, Seetin MG et al (2016) DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature 532:329–333
Xiong J, Ye TT, Ma CJ, Cheng QY, Yuan BF, Feng YQ (2019) N 6-hydroxymethyladenine: a hydroxylation derivative of N6-methyladenine in genomic DNA of mammals. Nucleic Acids Res 47:1268–1277
Gillingham D, Shahid R (2015) Catalysts for RNA and DNA modification. Curr Opin Chem Biol 25:110–114
Wu Y, Zhou C, Yuan Q (2018) Role of DNA and RNA N6-adenine methylation in regulating stem cell fate. Curr Stem Cell Res Ther 13:31–38
Zhang C, Jia G (2018) Reversible RNA modification N(1)-methyladenosine (m(1)A) in mRNA and tRNA. Genom Proteom Bioinform 16:155–161
Zhou C, Liu Y, Li X, Zou J, Zou S (2016) DNA N(6)-methyladenine demethylase ALKBH1 enhances osteogenic differentiation of human MSCs. Bone Res 4:16033
Wang C, Huang Y, Zhang J, Fang Y (2020) MiRNA-339–5p suppresses the malignant development of gastric cancer via targeting ALKBH1. Exp Mol Pathol 115:104449
Liu Y, Yuan Q, Xie L (2018) The AlkB family of Fe (II)/alpha-ketoglutarate-dependent dioxygenases modulates embryogenesis through epigenetic regulation. Curr Stem Cell Res Ther 13:136–143
Zhou Z, Li HQ, Liu F (2018) DNA methyltransferase inhibitors and their therapeutic potential. Curr Top Med Chem 18:2448–2457
Roy TW, Bhagwat AS (2007) Kinetic studies of Escherichia coli AlkB using a new fluorescence-based assay for DNA demethylation. Nucleic Acids Res 35:e147
Widagdo J, Anggono V (2018) The m6A-epitranscriptomic signature in neurobiology: from neurodevelopment to brain plasticity. J Neurochem 147:137–152
Muller TA, Struble SL, Meek K, Hausinger RP (2018) Characterization of human AlkB homolog 1 produced in mammalian cells and demonstration of mitochondrial dysfunction in ALKBH1-deficient cells. Biochem Biophys Res Commun 495:98–103
Weng H, Huang H, Wu H et al (2018) METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell 22:191-205 e199
Li M, Zhao X, Wang W et al (2018) Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol 19:69
Paris J, Morgan M, Campos J et al (2019) Targeting the RNA m(6)A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell 25:137–148 e136
Wakisaka KT, Muraoka Y, Shimizu J et al (2019) Drosophila alpha-ketoglutarate-dependent dioxygenase AlkB is involved in repair from neuronal disorders induced by ultraviolet damage. NeuroReport 30:1039–1047
Muller TA, Tobar MA, Perian MN, Hausinger RP (2017) Biochemical characterization of AP lyase and m(6)A demethylase activities of human AlkB homologue 1 (ALKBH1). Biochemistry 56:1899–1910
Valastyan S, Weinberg RA (2011) Tumor metastasis: molecular insights and evolving paradigms. Cell 147:275–292
Ougland R, Jonson I, Moen MN et al (2016) Role of ALKBH1 in the core transcriptional network of embryonic stem cells. Cell Physiol Biochem 38:173–184
Welford RW, Schlemminger I, McNeill LA, Hewitson KS, Schofield CJ (2003) The selectivity and inhibition of AlkB. J Biol Chem 278:10157–10161
Toh JDW, Sun L, Lau LZM et al (2015) A strategy based on nucleotide specificity leads to a subfamily-selective and cell-active inhibitor of N(6)-methyladenosine demethylase FTO. Chem Sci 6:112–122
Bian K, Chen F, Humulock ZT, Tang Q, Li D (2017) Copper inhibits the AlkB family DNA repair enzymes under Wilson’s disease condition. Chem Res Toxicol 30:1794–1796
Xie LJ, Liu L, Cheng L (2020) Selective inhibitors of AlkB family of nucleic acid demethylases. Biochemistry 59:230–239
Das M, Yang T, Dong J et al (2018) Multiprotein dynamic combinatorial chemistry: a strategy for the simultaneous discovery of subfamily-selective inhibitors for nucleic acid demethylases FTO and ALKBH3. Chem Asian J 13:2854–2867
Chen Y, Hong T, Wang S, Mo J, Tian T, Zhou X (2017) Epigenetic modification of nucleic acids: from basic studies to medical applications. Chem Soc Rev 46:2844–2872
Acknowledgements
This work was supported by the Youth Scientific Research Training Project of GZUCM (No. 2019QNPY07), the National Natural Science Foundation of China (No. 31700625), Guangdong Key Laboratory for Translational Cancer Research of Chinese Medicine (No. 2018B030322011), Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (Caiyan Wang, 2019). In addition, we also thanked three authors, who participated in the revision of manuscript. Zibo Li contributed to the revision of paper, Lin Jiang and Liangkai Cheng prepared the pictures for the revised manuscript.
Author information
Authors and Affiliations
Contributions
CW conceived the project and designed the review; YZ completed research literature analysis literature experimental data, and wrote the paper; CW and YZ together advised and revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, Y., Wang, C. Demethyltransferase AlkBH1 substrate diversity and relationship to human diseases. Mol Biol Rep 48, 4747–4756 (2021). https://doi.org/10.1007/s11033-021-06421-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06421-x