Abstract
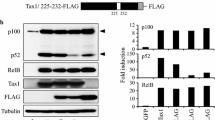
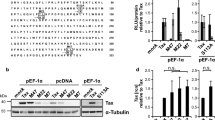
Transcription factor Ets-2 downregulates the expression of cytokine genes and HIV-1 in resting T-cells. Herein, we studied whether Ets-2 regulates the expression of lymphotropic factors (LFs) NFAT2, NF-κΒ/p65, c-Jun, c-Fos, which regulate the activation/differentiation of T-cells, and kinase CDK10, which controls Ets-2 degradation and repression activity. In silico analysis revealed Ets-2 binding sites on the promoters of NFAT2, c-Jun, c-Fos. The T-cell lines Jurkat (models T-cell signaling/activation) and H938 (contains the HIV-1-LTR) were transfected with an Ets-2 overexpressing vector, in the presence/absence of mitogens. mRNA and protein levels were assessed by qPCR and Western immunoblotting, respectively. Ets-2 overexpression in unstimulated Jurkat increased NFAT2 and c-Jun mRNA/protein, c-Fos mRNA and NF-κΒ/p65 protein, and decreased CDK10 protein. In unstimulated H938, Ets-2 upregulated NFAT2, c-Jun and CDK10 mRNA/protein and NF-κΒ/p65 protein. In stimulated Jurkat, Ets-2 increased NFAT2, c-Jun and c-Fos mRNA/protein and decreased CDK10 mRNA/protein. In stimulated H938 Ets-2 increased NFAT2, c-Jun and c-Fos protein and reduced CDK10 protein levels. Furthermore, Ets-2 overexpression modulated the expression of pro- and anti-apoptotic genes in both cell lines. Ets-2 upregulates the expression of key LFs involved in the activation of cytokine genes or HIV-1 in T-cells, either through its physical interaction with gene promoters or through its involvement in signaling pathways that directly impact their expression. The effect of Ets-2 on CDK10 expression in H938 vs Jurkat cells dictates that, additionally to Ets-2 degradation, CDK10 may facilitate Ets-2 repression activity in cells carrying the HIV-1-LTR, contributing thus to the regulation of HIV latency in virus-infected T-cells.






Similar content being viewed by others
References
Anderson MK, Hernandez-Hoyos G, Diamond RA, Rothenberg EV (1999) Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development 126:3131–3148
Dwyer JM, Liu JP (2010) Ets2 transcription factor, telomerase activity and breast cancer. Clin Exp Pharmacol Physiol 37:83–87. https://doi.org/10.1111/j.1440-1681.2009.05236.x
Fry EA, Inoue K (2018) Aberrant expression of ETS1 and ETS2 proteins in cancer. Cancer Rep Rev 2:10. https://doi.org/10.15761/CRR.1000151
Wei GH, Badis G, Berger MF et al (2010) Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. Embo J 29:2147–2160. https://doi.org/10.1038/emboj.2010.106
Hsu T, Trojanowska M, Watson DK (2004) Ets proteins in biological control and cancer. J Cell Biochem 91:896–903. https://doi.org/10.1002/jcb.20012
Selvaraj N, Kedage V, Hollenhorst PC (2015) Comparison of MAPK specificity across the ETS transcription factor family identifies a high-affinity ERK interaction required for ERG function in prostate cells. Cell Commun Signal 13:015–0089. https://doi.org/10.1186/s12964-015-0089-7
Zaldumbide A, Carlotti F, Pognonec P, Boulukos KE (2002) The role of the Ets2 transcription factor in the proliferation, maturation, and survival of mouse thymocytes. J Immunol 169:4873–4881. https://doi.org/10.4049/jimmunol.169.9.4873
Yamamoto H, Flannery ML, Kupriyanov S, Pearce J, McKercher SR, Henkel GW et al (1998) Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev 12:1315–1326. https://doi.org/10.1101/gad.12.9.1315
Oikawa T (2004) ETS transcription factors: possible targets for cancer therapy. Cancer Sci 95:626–633. https://doi.org/10.1111/j.1349-7006.2004.tb03320.x
Mouzaki A, Rungger D (1994) Properties of transcription factors regulating interleukin-2 gene transcription through the NFAT binding site in untreated or drug-treated naive and memory T-helper cells. Blood 84:2612–2621. https://doi.org/10.1182/blood.V84.8.2612.2612
Marson A, Kretschmer K, Frampton GM et al (2007) Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature 445:931–935. https://doi.org/10.1038/nature05478
Argyropoulos C, Nikiforidis GC, Theodoropoulou M et al (2004) Mining microarray data to identify transcription factors expressed in naive resting but not activated T lymphocytes. Genes Immun 5:16–25. https://doi.org/10.1038/sj.gene.6364034
Bunting K, Wang J, Shannon MF (2006) Control of interleukin-2 gene transcription: a paradigm for inducible, tissue-specific gene expression. Vitam Horm 74:105–145. https://doi.org/10.1016/S0083-6729(06)74005-5
Panagoulias I, Georgakopoulos T, Aggeletopoulou I, Agelopoulos M, Thanos D, Mouzaki A (2016) Transcription factor Ets-2 acts as a preinduction repressor of interleukin-2 (IL-2) transcription in naive T helper lymphocytes. J Biol Chem 291:26707–26721. https://doi.org/10.1074/jbc.M116.762179
Toumpeki C, Anastasakis D, Panagoulias I et al (2018) Construction of an M1GS ribozyme for targeted and rapid mRNA cleavage; application on the Ets-2 oncogene. Med Chem 14:604–616. https://doi.org/10.2174/1573406414666180112115201
Panagoulias I, Karagiannis F, Aggeletopoulou I et al (2018) Ets-2 acts as a transcriptional repressor of the human immunodeficiency virus type 1 through binding to a repressor-activator target sequence of 5′-LTR. Front Immunol 8:1924. https://doi.org/10.3389/fimmu.2017.01924
Basuyaux JP, Ferreira E, Stéhelin D, Butticè G (1997) The Ets transcription factors interact with each other and with the c-Fos/c-Jun complex via distinct protein domains in a DNA-dependent and -independent manner. J Biol Chem 272:26188–26195. https://doi.org/10.1074/jbc.272.42.26188
Felber BK, Pavlakis GN (1988) A quantitative bioassay for HIV-1 based on trans-activation. Science 239:184–187. https://doi.org/10.1126/science.3422113
Schwartz S, Felber BK, Fenyo EM, Pavlakis GN (1989) Rapidly and slowly replicating human immunodeficiency virus type 1 isolates can be distinguished according to target-cell tropism in T-cell and monocyte cell lines. Proc Natl Acad Sci USA 86:7200–7203. https://doi.org/10.1073/pnas.86.18.7200
Kreft L, Soete A, Hulpiau P, Botzki A, Saeys Y, De Bleser P (2017) ConTrav3: a tool to identify transcription factor binding sites across species, update 2017. Nucleic Acids Res 45:W490–W494. https://doi.org/10.1093/nar/gkx376
Macian F (2005) NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol 5:472–484. https://doi.org/10.1038/nri1632
Gazon H, Barbeau B, Mesnard JM, Peloponese JM Jr (2018) Hijacking of the AP-1 signaling pathway during development of ATL. Front Microbiol 8:2686. https://doi.org/10.3389/fmicb.2017.02686
Liu T, Zhang L, Joo D, Sun SC (2017) NF-κB signaling in inflammation. Signal Transduct Target Ther 2:17023. https://doi.org/10.1038/sigtrans.2017.23
Liu X, Berry CT, Ruthel G et al (2016) T cell receptor-induced nuclear factor κB (NF-κB) signaling and transcriptional activation are regulated by STIM1- and orai1-mediated calcium entry. J Biol Chem 291:8440–8452. https://doi.org/10.1074/jbc.M115.713008
Guen VJ, Gamble C, Lees JA, Colas P (2017) The awakening of the CDK10/Cyclin M protein kinase. Oncotarget 8:50174–50186. https://doi.org/10.18632/oncotarget.15024
Carrero ZI, Kollareddy M, Chauhan KM, Ramakrishnan G, Martinez LA (2016) Mutant p53 protects ETS2 from non-canonical COP1/DET1 dependent degradation. Oncotarget 7:12554–12567. https://doi.org/10.18632/oncotarget.7275
Kasten M, Giordano A (2001) Cdk10, a Cdc2-related kinase, associates with the Ets2 transcription factor and modulates its transactivation activity. Oncogene 20:1832–1838. https://doi.org/10.1038/sj.onc.1204295
Montano M (2014) Translational biology in medicine. Woodhead publishing series in biomedicine. Woodhead Publishing, Oxford, pp 9–33
Pessler F, Cron R (2004) Reciprocal regulation of the nuclear factor of activated T cells and HIV-1. Genes Immun 5:158–167. https://doi.org/10.1038/sj.gene.6364047
Argyropoulos C, Mouzaki A (2006) Immunosuppressive drugs in HIV disease. Curr Top Med Chem 6:1769–1789. https://doi.org/10.2174/156802606778194271
Milanovic M, Kracht M, Schmitz ML (2014) The cytokine-induced conformational switch of nuclear factor κB p65 is mediated by p65 phosphorylation. Biochem J 457:401–413. https://doi.org/10.1042/BJ20130780
Christian F, Smith EL, Carmody RJ (2016) The regulation of NF-κB subunits by phosphorylation. Cells 5:12. https://doi.org/10.3390/cells5010012
Hurd TW, Culbert AA, Webster KJ, Tavaré JM (2002) Dual role for mitogen-activated protein kinase (Erk) in insulin-dependent regulation of Fra-1 (fos-related antigen-1) transcription and phosphorylation. Biochem J 368(Pt 2):573–580. https://doi.org/10.1042/BJ20020579
Rosenberger SF, Finch JS, Gupta A, Bowden GT (1999) Extracellular signal-regulated kinase 1/2-mediated phosphorylation of JunD and FosB is required for okadaic acid-induced activator protein 1 activation. J Biol Chem 274(2):1124–1130
Adler J, Reuven N, Kahana C, Shaul Y (2010) c-Fos proteasomal degradation is activated by a default mechanism, and its regulation by NAD(P)H:quinone oxidoreductase 1 determines c-Fos serum response kinetics. Mol Cell Biol 30(15):3767–3778. https://doi.org/10.1128/MCB.00899-09
Iorns E, Turner NC, Elliott R et al (2008) Identification of CDK10 as an important determinant of resistance to endocrine therapy for breast cancer. Cancer Cell 13:91–104. https://doi.org/10.1016/j.ccr.2008.01.001
Wolvetang EJ, Wilson TJ, Sanij E et al (2003) ETS2 overexpression in transgenic models and in Down syndrome predisposes to apoptosis via the p53 pathway. Hum Mol Genet 12:247–255. https://doi.org/10.1093/hmg/ddg015
van Montfort T, van der Sluis R, Darcis G et al (2019) Dendritic cells potently purge latent HIV-1 beyond TCR-stimulation, activating the PI3K-Akt-mTOR pathway. EBioMedicine 42:97–108. https://doi.org/10.1016/j.ebiom.2019.02.014
Funding
This research was co-financed by Greece and the European Union (European Social Fund- ESF) through the Operational Program “Human Resources Development, Education and Lifelong Learning” in the context of the project “Strengthening Human Resources Research Potential via Doctorate Research” (MIS-5000432), implemented by the State Scholarships Foundation (ΙΚΥ) (to PD), the Operational Program «Human Resources Development, Education and Lifelong Learning» in the context of the project "Reinforcement of Postdoctoral Researchers—2nd Cycle" (MIS-5033021), implemented by the State Scholarships Foundation (ΙΚΥ) (to IA), and GILEAD-ASKLEPIOS grants for 2018 and 2019 (to AM).
Author information
Authors and Affiliations
Contributions
AM conceived and coordinated the study; PD, IA, IP performed the experiments, analyzed the data and prepared the figures; TG and IP advised on methodology; PD, IA, TG and AM wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
None to declare.
Consent for publication
The manuscript has been read and approved by all named authors and there are no other persons who satisfied the criteria for authorship but are not listed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Davoulou, P., Aggeletopoulou, I., Panagoulias, I. et al. Transcription factor Ets-2 regulates the expression of key lymphotropic factors. Mol Biol Rep 47, 7871–7881 (2020). https://doi.org/10.1007/s11033-020-05865-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05865-x