Abstract
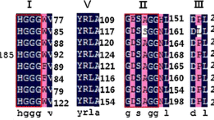
A functional screening of 1152 clones from a plasmid library constructed with DNA extracted from Brazilian mangrove sediments revealed 3 positive clones for ester-hydrolyzing enzymes, or about one lipolytic gene per 1.2 Mb DNA, which corroborates the idea that oil-contaminated mangroves are a good source of novel microbial lipases/esterases. The partial sequence of the clone LipG7 (1179 bp) showed 30.2% of predicted structure identity with a known esterase, confirming LipG7 as a new member of family VIII esterases. LigG7 shared 80% sequence identity with 1,4-butanediol diacrylate esterase from the Gammaprotebacteria Porticoccus hydrocarbonoclasticus, suggesting it belongs to the Porticoccaceae family. LipG7 was heterologously expressed in Escherichia coli Rosetta-Gami DE3; the purified recombinant enzyme exhibited a predicted molecular weight of 45.2 kDa and exceptional activity towards 4-nitrophenyl butyrate, compared with other recombinant esterases, highlighting its enormous potential for biological applications.









Similar content being viewed by others
References
Choi JM, Han SS, Kim HS (2015) Industrial applications of enzyme biocatalysis: current status and future aspects. Biotechnol Adv 33:1443–1454. https://doi.org/10.1016/j.biotechadv.2015.02.014
Rappé MS, Giovannoni SJ (2003) The uncultured microbial majority. Annu Rev Microbiol 57:369–394. https://doi.org/10.1146/annurev.micro.57.030502.090759
Handelsman J (2004) Metagenomics : application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev 68:669–685. https://doi.org/10.1128/MBR.68.4.669
Kim H, Lee K, Kim M, Lee J, Kim K (2018) Identification and characterization of a novel KG42 xylanase (GH10 family) isolated from the black goat rumen-derived metagenomic library. Carbohydr Res 469:1–9. https://doi.org/10.1016/j.carres.2018.08.010
Ottoni JR (2018) Functional metagenomics of oil-impacted mangrove sediments reveals high abundance of hydrolases of biotechnological interest. World J Microbiol Biotechnol 33:1–13. https://doi.org/10.1007/s11274-017-2307-5
Song Y, Lee K, Baek J, Kim M, Kwon M, Kim Y, Park M, Ko H, Lee J, Kim K (2017) Biotechnology and Industrial Microbiology Isolation and characterization of a novel endo-β-1,4-glucanase from a metagenomic library of the black-goat rumen. Braz J Microbiol 8:801–808. https://doi.org/10.1016/j.bjm.2017.03.006
Gao W, Wu K, Chen L, Fan H, Zhao Z, Gao B, Wang H, Wei D (2016) A novel esterase from a marine mud metagenomic library for biocatalytic synthesis of short-chain flavor esters. Microb Cell Fact 15:41. https://doi.org/10.1186/s12934-016-0435-5
Jaeger KE, Eggert T (2002) Lipases for biotechnology. Curr Opin Biotechnol 13:390–397. https://doi.org/10.1016/S0958-1669(02)00341-5
Mouad AM, Taupin D, Lehr L, Yvergnaux F, Porto ALM (2016) Aminolysis of linoleic and salicylic acid derivatives with Candida antarctica lipase B: a solvent-free process to obtain amphiphilic amides for cosmetic application. J Mol Catal B Enzym 126:64–68. https://doi.org/10.1016/j.molcatb.2016.01.002
Avhad MR, Marchetti JM (2019) Uses of enzymes for biodiesel production. In: Advanced bioprocessing for alternative fuels, biobased chemicals, and bioproducts. Woodhead Publishing, Sawston, pp 135–152
Sarmah N, Revathi D, Sheelu G, Yamuna Rani K, Sridhar S, Mehtab V, Sumana C (2018) Recent advances on sources and industrial applications of lipases. Biotechnol Prog 34:5–28. https://doi.org/10.1002/btpr.2581
Arpigny JL, Jaeger K-E (1999) Bacterial lipolytic enzymes: classification and properties. Biochem J 343:177–183. https://doi.org/10.1042/bj3430177
Gupta R, Kumari A, Syal P, Singh Y (2015) Molecular and functional diversity of yeast and fungal lipases: their role in biotechnology and cellular physiology. Prog Lipid Res 57:40–54. https://doi.org/10.1016/j.plipres.2014.12.001
BBC Research (2018) Global markets for enzymes in industrial applications, BBC Research Report
Holguin G, Gonzalez-Zamorano P, De-Bashan LE, Mendoza R, Amador E, Bashan Y (2006) Mangrove health in an arid environment encroached by urban development—a case study. Sci Total Environ 363:260–274. https://doi.org/10.1016/j.scitotenv.2005.05.026
Couto GH, Glogauer A, Faoro H, Chubatsu LS, Souza EM, Pedrosa FO (2010) Isolation of a novel lipase from a metagenomic library derived from mangrove sediment from the south Brazilian coast. Genet Mol Res 9:514–523. https://doi.org/10.4238/vol9-1gmr738
Castro RA, Quecine M, Lacava PT, Batista BD, Luvizotto DM, Marcon J, Ferreira A, Melo IS, Azevedo JL (2014) Isolation and enzyme bioprospection of endophytic bacteria associated with plants of Brazilian mangrove ecosystem. SpringerPlus 3:382. https://doi.org/10.1186/2193-1801-3-382
Uchiyama T, Abe T, Ikemura T, Watanabe K (2005) Substrate-induced gene-expression screening of environmental metagenome libraries for isolation of catabolic genes. Nat Biotechnol 23:88–93. https://doi.org/10.1038/nbt1048
Birnboim HC (1983) A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol 100:243–255. https://doi.org/10.1016/0076-6879(83)00059-2
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. https://doi.org/10.1093/nar/25.17.3389
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. https://doi.org/10.1093/nar/22.22.4673
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526. https://doi.org/10.1093/oxfordjournals.molbev.a040023
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31:3381–3385. https://doi.org/10.1093/nar/gkg520
Sambrook J (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Knox JR, Moews PC, Frere JM (1996) Molecular evolution of bacterial beta-lactam resistance. Chem Biol 3:937–947. https://doi.org/10.1016/s1074-5521(96)90182-9
Lee HW, Jung WK, Kim YH, Ryu BH, Kim TD, Kim J, Kim H (2016) Characterization of a novel alkaline family VIII esterase with S-enantiomer preference from a compost metagenomic library. J Microbiol Biotechnol 26(2):315–325. https://doi.org/10.4014/jmb.1509.09081
Jiménez-Osés G, Osuna S, Gao X, Sawaya MR, Gilson L, Collier SJ, Huisman GW, Yeates TO, Tang Y, Houk KN (2014) The role of distant mutations and allosteric regulation on LovD active site dynamics. Nat Chem Biol 10:431. https://doi.org/10.1038/nchembio.1503
Gutierrez T, Nichols PD, Whitman WB, Aitken MD (2012) Porticoccus hydrocarbonoclasticus sp. nov., an aromatic hydrocarbon-degrading bacterium identified in laboratory cultures of marine phytoplankton. Appl Environ Microbiol 78:628–637. https://doi.org/10.1128/AEM.06398-11
Oh HM, Kim H, Kim KM, Min GS, Cho JC (2010) Porticoccus litoralis gen. nov., sp. nov., a gammaproteobacterium isolated from the Yellow Sea. Int J Syst Evol Microbiol 60:727–732. https://doi.org/10.1099/ijs.0.013938-0
Ribicic D, Netzer R, Hazen TC, Techtmann SM, Drabløs F, Brakstad OG (2018) Microbial community and metagenome dynamics during biodegradation of dispersed oil reveals potential key-players in cold Norwegian seawater. Mar Pollut Bull 129:370–378. https://doi.org/10.1016/j.marpolbul.2018.02.034
Lam KN, Cheng J, Engel K, Neufeld JD, Charles TC (2015) Current and future resources for functional metagenomics. Front Microbiol 6:1–8. https://doi.org/10.3389/fmicb.2015.01196
Henne A, Schmitz RA, Bömeke M, Gottschalk G, Daniel R (2000) Screening of environmental DNA libraries for the presence of genes conferring lipolytic activity on Escherichia coli. Appl Environ Microbiol 66(7):3113–3116. https://doi.org/10.1128/aem.66.7.3113-3116.2000
Pereira MR, Mercaldi GF, Maester TC, Balan A, Lemos EGM (2015) Est16, a new esterase isolated from a metagenomic library of a microbial consortium specializing in diesel oil degradation. PLoS ONE 10:1–16. https://doi.org/10.1371/journal.pone.0133723
Andrade MVF, Santos FR, Oliveira AHB, Nascimento RF, Cavalcante RM (2019) Influence of sediment parameters on the distribution and fate of PAHs in an estuarine tropical region located in the Brazilian semi-arid (Jaguaribe River, Ceará coast). Mar Pollut Bull 146:703–710. https://doi.org/10.1016/j.marpolbul.2019.07.027
Oliveira AHB, Cavalcante RM, Duaví WC, Fernandes GM, Nascimento RF, Queiroz MELR, Mendonça KV (2016) The legacy of organochlorine pesticide usage in a tropical semi-arid region (Jaguaribe River, Ceará, Brazil): implications of the influence of sediment parameters on occurrence, distribution and fate. Sci Total Environ 542:254–263. https://doi.org/10.1016/j.scitotenv.2015.10.058
Kovacic F, Babic N, Krauss U, Jaeger K (2019) Classification of lipolytic enzymes from bacteria. Aerobic utilization of hydrocarbons, oils and lipids. Springer, Cham. https://doi.org/10.1007/978-3-319-39782-5
Williams JD (1999) Beta-lactamases and beta-lactamase inhibitors. Int J Antimicrob Agents 12(Suppl 1):S3–S7; discussion S26–S7. https://doi.org/10.1016/s0924-8579(99)00085-0
Hitch TCA, Clavel T (2019) A proposed update for the classification and description of bacterial lipolytic enzymes. PeerJ 7:e7249. https://doi.org/10.7717/peerj.7249
Rashamuse K, Magomani V, Ronneburg T, Brady D (2009) A novel family VIII carboxylesterase derived from a leachate metagenome library exhibits promiscuous B-lactamase activity on nitrocefin. Appl Microbiol Biotechnol 83:491–500. https://doi.org/10.1007/s00253-009-1895-x
Li X, Guo J, Hu Y, Yang Y (2019) Identification of a novel feruloyl esterase by functional screening of a soil metagenomic library. Appl Biochem Biotechnol 187:424–437. https://doi.org/10.1007/s12010-018-2832-1
Zhu Y, Li J, Cai H, Ni H, Xiao A, Hou L (2013) Characterization of a new and thermostable esterase from a metagenomic library. Microbiol Res 168:589–597. https://doi.org/10.1016/j.micres.2013.04.004
Kim YH, Kwon EJ, Kim SK, Jeong YS, Kim J, Yun HD, Kim H (2010) Molecular cloning and characterization of a novel family VIII alkaline esterase from a compost metagenomic library. Biochem Biophys Res Commun 393:45–49. https://doi.org/10.1016/j.bbrc.2010.01.070
Choi YJ, Miguez CB, Lee BH (2004) Characterization and heterologous gene expression of a novel esterase from Lactobacillus casei CL96. Society 70:3213–3221. https://doi.org/10.1128/AEM.70.6.3213
Voget S, Leggewie C, Uesbeck A, Raasch C, Jaeger KE, Streit WR (2003) Prospecting for novel biocatalysts in a soil metagenome. Appl Environ Microbiol 69:6235–6242. https://doi.org/10.1128/AEM.69.10.6235-6242.2003
Acknowledgements
The authors acknowledge the contributions made by the blinded reviewers and are grateful to Thomas Hitch (Institute of Medical Microbiology, Aachen, Germany) for the help with the enzyme classification analysis. The authors thank the National Council for Scientific and Technological Development (CNPq) and Foundation for Scientific and Technological Development of Ceará (FUNCAP) for the grant provided to LRBG (PRONEX-PR2-0101-00012.01.00/15) and for the scholarship provided to FJA. Collection of samples was registered by the pertinent environmental agency (A982E7D-SisGen).
Funding
The authors thank the National Council for Scientific and Technological Development (CNPq) and Foundation for Scientific and Technological Development of Ceará (FUNCAP) for the grant provided to LRBG (PRONEX-PR2-0101–00012.01.00/15) and for the scholarship provided to FJA.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Francisco J. Araujo, Gabrielly O. Silva and Vanessa L.R Nogueira. Luciana R. B. Gonçalves has contributed with funding acquisition and resources. Formal analysis and investigation were performed by Denise C. Hissa and Vânia M. M. Melo. The first draft of the manuscript was written by Denise C. Hissa and André S. L. M. Antunes. Review and editing of the manuscript was performed by Vânia M. M. Melo. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declared that they have no conflict of interest.
Ethics approval
Collection of samples was registered by the pertinent environmental agency (A982E7D-SisGen).
Informed consent
All co-authors have agreed to participate. All co-authors have agreed for this manuscript publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Araujo, F.J., Hissa, D.C., Silva, G.O. et al. A novel bacterial carboxylesterase identified in a metagenome derived-clone from Brazilian mangrove sediments. Mol Biol Rep 47, 3919–3928 (2020). https://doi.org/10.1007/s11033-020-05484-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05484-6