Abstract
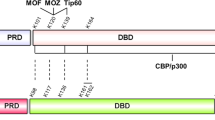
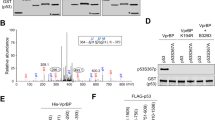
As a “genomic guardian”, p53 mainly functions as a transcription factor that regulates downstream targets responsible for cell fate control, and the activity of p53 is tightly regulated by a complex network that include an abundance of post-translational modifications. Notably, acetylation of p53 at many positions has been demonstrated to play a major role in accurate p53 regulation and cell fate determination. However, no evidence has been provided to compare the effect of acetylation at different sites on p53 regulation. Here, we constructed six acetylation-defective p53 mutants that lysine was substituted by arginine at residues 120, 164, 305, 320, 370/372/373 or 381/382/386, respectively, and determined their effects on p53 activity systematically. Our results showed that all six mutants exhibited diminished transactivation ability and selective regulation of target genes expression through distinct mechanisms. Specifically, lysine 370/372/373 and 381/382/386 mutations decreased p53 stability, and lysine 305 mutation reduced p53 phosphorylation level at serine 15, while lysine 120 and 164 mutations decreased p53 acetylation level at lysine 382. Collectively, these data indicate that acetylation of p53 at different sites has diverse regulatory effects on p53 transcriptional activity through different mechanisms.





Similar content being viewed by others
Abbreviations
- PTM:
-
Post-translational modifications
- HATs:
-
Histone/lysine acetyltransferases
- CTD:
-
C-terminal domain
- PCAF:
-
p300/CBP associated factor
- K120:
-
Lysine 120
- K164:
-
Lysine 164
- K305:
-
Lysine 305
- K320:
-
Lysine 320
- K382/383:
-
Lysine 382/383
- S15:
-
Serine 15
- pG13-Luc:
-
pG13L containing 13 tandem p53 binding site repeats
- K120R:
-
Mutants with lysine to arginine changes at residues 120
- K164R:
-
Mutants with lysine to arginine changes at residues 164
- K305R:
-
Mutants with lysine to arginine changes at residues 305
- K320R:
-
Mutants with lysine to arginine changes at residues 320
- K370/372/373R:
-
Mutants with lysine to arginine changes at residues 370/372/373
- K381/382/386R:
-
Mutants with lysine to arginine changes at residues 381/382/386
- CHX:
-
Cycloheximide
References
Kastenhuber ER, Lowe SW (2017) Putting p53 in context. Cell 170:2017
Mello SS, Attardi LD (2018) Deciphering p53 signaling in tumor suppression. Curr Opin Cell Biol 51:2018
Kruse JP, Gu W (2009) Modes of p53 regulation. Cell 137:2009
Gu B, Zhu WG (2012) Surf the post-translational modification network of p53 regulation. Int J Biol Sci 8:2012
Reed SM, Quelle DE (2014) p53 Acetylation: regulation and consequences. Cancers 7:2014
Zhu WG (2017) Regulation of p53 acetylation. Science China. Life Sci 60:2017
Wang SJ, Li D, Ou Y, Jiang L, Chen Y, Zhao Y, Gu W (2016) Acetylation is crucial for p53-mediated ferroptosis and tumor suppression. Cell Rep 17:2016
Wang D, Kon N, Lasso G, Jiang L, Leng W, Zhu WG, Qin J, Honig B, Gu W (2016) Acetylation-regulated interaction between p53 and SET reveals a widespread regulatory mode. Nature 538:2016
Gu W, Roeder RG (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:1997
Feng L, Lin T, Uranishi H, Gu W, Xu Y (2005) Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol Cell Biol 25:2005
Zhao Y, Lu S, Wu L, Chai G, Wang H, Chen Y, Sun J, Yu Y, Zhou W, Zheng Q, Wu M, Otterson GA, Zhu WG (2006) Acetylation of p53 at lysine 373/382 by the histone deacetylase inhibitor depsipeptide induces expression of p21(Waf1/Cip1). Mol Cell Biol 26:2006
Tang Y, Zhao W, Chen Y, Zhao Y, Gu W (2008) Acetylation is indispensable for p53 activation. Cell 133:2008
Wang YH, Tsay YG, Tan BC, Lo WY, Lee SC (2003) Identification and characterization of a novel p300-mediated p53 acetylation site, lysine 305. J Biol Chem 278:2003
Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL (2001) Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell 8:2001
Knights CD, Catania J, Di Giovanni S, Muratoglu S, Perez R, Swartzbeck A, Quong AA, Zhang X, Beerman T, Pestell RG, Avantaggiati ML (2006) Distinct p53 acetylation cassettes differentially influence gene-expression patterns and cell fate. J Cell Biol 173:2006
Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB (2006) Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell 24:2006
Tang Y, Luo J, Zhang W, Gu W (2006) Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell 24:2006
Wang S, Tian C, Xiao T, Xing G, He F, Zhang L, Chen H (2010) Differential regulation of Apak by various DNA damage signals. Mol Cell Biochem 333:2010
Tian C, Xing G, Xie P, Lu K, Nie J, Wang J, Li L, Gao M, Zhang L, He F (2009) KRAB-type zinc-finger protein Apak specifically regulates p53-dependent apoptosis. Nat Cell Biol 11:2009
Wang SJ, Gu W (2014) To be, or not to be: functional dilemma of p53 metabolic regulation. Curr Opin Oncol 26:2014
Menendez D, Inga A, Resnick MA (2009) The expanding universe of p53 targets. Nat Rev Cancer 9:2009
Shu KX, Li B, Wu LX (2007) The p53 network: p53 and its downstream genes. Colloids Surf B 55:2007
Li M, Luo J, Brooks CL, Gu W (2002) Acetylation of p53 inhibits its ubiquitination by Mdm2. J Biol Chem 277:2002
Ito A, Kawaguchi Y, Lai CH, Kovacs JJ, Higashimoto Y, Appella E, Yao TP (2002) MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J 21:2002
Meulmeester E, Pereg Y, Shiloh Y, Jochemsen AG (2005) ATM-mediated phosphorylations inhibit Mdmx/Mdm2 stabilization by HAUSP in favor of p53 activation. Cell Cycle 4:2005
Loughery J, Cox M, Smith LM, Meek DW (2014) Critical role for p53-serine 15 phosphorylation in stimulating transactivation at p53-responsive promoters. Nucleic Acids Res 42:2014
Chao C, Wu Z, Mazur SJ, Borges H, Rossi M, Lin T, Wang JY, Anderson CW, Appella E, Xu Y (2006) Acetylation of mouse p53 at lysine 317 negatively regulates p53 apoptotic activities after DNA damage. Mol Cell Biol 26:2006
Rodriguez MS, Desterro JM, Lain S, Lane DP, Hay RT (2000) Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol Cell Biol 20:2000
Nakamura S, Roth JA, Mukhopadhyay T (2000) Multiple lysine mutations in the C-terminal domain of p53 interfere with MDM2-dependent protein degradation and ubiquitination. Mol Cell Biol 20:2000
Ou YH, Chung PH, Sun TP, Shieh SY (2005) p53 C-terminal phosphorylation by CHK1 and CHK2 participates in the regulation of DNA-damage-induced C-terminal acetylation. Mol Biol Cell 16:2005
Jenkins LM, Yamaguchi H, Hayashi R, Cherry S, Tropea JE, Miller M, Wlodawer A, Appella E, Mazur SJ (2009) Two distinct motifs within the p53 transactivation domain bind to the Taz2 domain of p300 and are differentially affected by phosphorylation. Biochemistry 48:2009
Lee CW, Ferreon JC, Ferreon AC, Arai M, Wright PE (2010) Graded enhancement of p53 binding to CREB-binding protein (CBP) by multisite phosphorylation. Proc Natl Acad Sci USA 107:2010
Lau AW, Liu P, Inuzuka H, Gao D (2014) SIRT1 phosphorylation by AMP-activated protein kinase regulates p53 acetylation. Am J Cancer Res 4:2014
Bode AM, Dong Z (2004) Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer 4:2004
Shahar OD, Gabizon R, Feine O, Alhadeff R, Ganoth A, Argaman L, Shimshoni E, Friedler A, Goldberg M (2013) Acetylation of lysine 382 and phosphorylation of serine 392 in p53 modulate the interaction between p53 and MDC1 in vitro. PLoS ONE 8:2013
Acknowledgements
We thank Dr. Bert Vogelstein (Johns Hopkins University) for providing the luciferase reporter plasmid pG13-Luc, Dr. Lingqiang Zhang (Beijing Institute of Lifeomics) for Human lung adenocarcinoma H1299 cell line.
Funding
This work was partially supported by grants from the National Natural Science Foundation Projects (31270799, 31771563), the National Key Research and Development Program of China (2017YFA0505700), the Chinese Program of International S&T Cooperation (2014DFB30020), and National Modern Agro (Dairy) Industry and Technology System (CARS-37).
Author information
Authors and Affiliations
Contributions
C.T. and J.W. conceived the project and designed the experiments. The experiments were performed by Y.W., Y.C., X.Z., Q.C., and H.W. Data were analyzed by C.T., J.W., Y.C. and Z.W. C.T. wrote the manuscript. Z.W. and J.W. provided critical proof reading of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, Y., Chen, Y., Chen, Q. et al. The role of acetylation sites in the regulation of p53 activity. Mol Biol Rep 47, 381–391 (2020). https://doi.org/10.1007/s11033-019-05141-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-019-05141-7