Abstract
Acinetobacter baumannii (A. baumannii) is an opportunistic pathogen that causes serious infections in the lungs, blood, and brain in critically ill hospital patients, resulting in considerable mortality rates every year. Due to the rapid appearance of multi-drug resistance or even pan-drug resistance isolates, it is becoming more and more difficult to cure A. baumannii infection by traditional antibiotic treatment, alternative strategies are urgently required to combat A. baumannii infection. In this study, we developed a DNA vaccine encoding two antigens from A. baumannii, OmpA and Pal, and the immunogenicity and protective efficacy was further evaluated. The results showed that the DNA vaccine exhibited significant immune protective efficacy against acute A. baumannii infection in a mouse pneumonia model, and cross protective efficacy was observed when immunized mice were challenged with clinical strains of A. baumannii. DNA vaccine immunization induced high level of humoral response and a mixed Th1/Th2/Th17 cellular response, which protect against lethal bacterial challenges by decreased bacterial loads and pathology in the lungs, and reduced level of inflammatory cytokines expression and inflammatory cell infiltration in BALF. These results demonstrated that it is possible to prevent A. baumannii infection by DNA vaccine and both OmpA and Pal could be serve as promising candidate antigens.
Similar content being viewed by others
Introduction
Acinetobacter baumannii (A. baumannii) is a Gram-negative opportunistic pathogen that has been identified as the etiological pathogen of severe nosocomial infections in patients, especially those with compromised immune systems and in the intensive care settings with several illness [1]. Infection caused by A. baumannii is mainly associated with but not limited to pneumonia, bacteremia, wound infections, urinary tract infections, and meningitis, resulting in considerable morbidity and mortality every year, the reported mortality associated with A. baumannii infection is up to 40% in the intensive care unit (ICU) settings, but varies largely between different infections and countries [2, 3]. Due to the rapid emergence of multi- and pan drug-resistant strains of A. baumannii, it has been classified as an ‘ESKAPE’ pathogen, which is a group of nosocomial infection pathogens with a high rate of antibiotic resistance and involve in nosocomial infections [4]. Meanwhile, A. baumannii is also identified as ‘Iraqibacter’ due to its sudden emergence in military treatment facilities during the Iraq War [1]. Antibiotic resistance has greatly decreased the effectiveness of antibiotic treatments, few antibiotics are effective for treating infections caused by this pathogen for the moment [5], and thus there is urgent requirement for the development of alternative therapeutic strategies to combat infection caused by A. baumannii.
Vaccination strategies are proved to be effective to prevent or treat infectious disease, and a series of researches have been focused on developing a vaccine against A. baumannii infection [6,7,8]. The antigens used in these studies including inactivated whole cells, cell components such as outer membrane vesicles (OMVs) and outer membrane complexes (OMCs), polysaccharide, and outer membrane proteins such as OmpA, Omp34 kDa and OprC [9,10,11,12]. Some of these antigens have been proven to be effective to protect against A. baumannii challenge in animal models, however, few of them entered clinical studies and no vaccine against A. baumannii was authorized for immunization in humans so far. This may be attributes to a series of reasons, for example, some of these antigens exhibited low immunogenicity and safety, some antigens showed limited protective efficacy, some of the protein antigens identified were outer membrane proteins and was difficult to express and purify.
DNA vaccines are proved to be efficient to provide protective immunity by direct injection of genetic material into a living host, they have potential advantages over conventional vaccines in preparation, stability, safety, and the ability to induce a wider range of immune response [13]. In this study, we constructed a bivalent DNA vaccine composed of plasmids pVAX1 encoding two outer membrane proteins from A. baumannii, OmpA and Pal. Among them, OmpA is regarded as the most promising antigen for the development of vaccines against A. baumannii infection, because it is a virulence factor that participated in the pathogenesis of A. baumannii and is highly immunogenic in animal models, immune with it alone was able to provide partial protection against lethal challenge of A. baumannii [14, 15]. Besides, OmpA is highly conserved among different strains and is the most abundant proteins identified in the OMVs isolated from A. baumannii [12]. Pal is a peptidoglycan-associated lipoprotein that present in the cell envelope and associated with the peptidoglycan layer, it contains an OmpA like domain in the C-terminal and plays an important role in the integrity of the outer membrane [16]. Almost all Pal proteins studied so far are highly immunogenic, which makes them attractive antigens for the production of subunit vaccines [17,18,19]. However, both proteins contain transmembrane domains and are difficult to express in E. coli, thus we developed a DNA vaccine based on the sequence of the two antigens, and the aim of this study was to evaluate the immunogenicity and protective efficacy of the bivalent DNA vaccine.
Materials and methods
Ethics statement
All animal care and use procedures in this study were in accordance with the Animal Ethics Procedures and Guide-lines of the People’s Republic of China. All animal experiments in this study were approved by the Animal Ethical and Experimental Committee of the Army Medical University (Chongqing, Permit No. 2011-04).
Bacterial strains and culture method
The A. baumannii LAC (Los Angeles County) -4 was kindly provided by Professor Chen [20]. Three clinical strains of A. baumannii (SJZ18, SJZ04 and SJZ28) were collected from Bethune International Peace Hospital (Hebei, China), the information of these strains was shown in Supplementary Table 1. All these strains were cultured in Tryptic Soy Broth at 37.0 °C overnight with constant shaking at 190 rpm, the bacterial suspension was centrifuged at 6000×g for 10 min, and the precipitate was washed with sterile PBS twice, and diluted to a suitable cell concentration determined spectrophotometrically at 600 nm (OD 600).
Mice
Six–eight week-old female C57BL/6 mice (weight at 18–22 grams) were purchased from Experimental Animal Center of Chongqing Medical University, under specific pathogen-free (SPF) condition.
Construction of DNA vaccines
The genomic DNA extracted from A. baumannii strain LAC-4 was used as template. The gene encoding OmpA and Pal (without signal peptide) was amplified from the genome DNA, and then inserted into an expression vector derived from the pVAX1 plasmid (Trans Gen biotech Company, China) and placed between BamHI and XhoI restriction sites, respectively. The plasmid of pVAX1-OmpA-Pal was synthesized by Sangon (Shanghai, China), a DNA sequence encoding the “GGGGS” linker was placed between the sequence of OmpA and Pal. All plasmids were confirmed by restriction endonuclease digestion and DNA sequencing. The recombinant plasmids (pVAX1-OmpA, pVAX1-Pal and pVAX1-OmpA-Pal) were transformed into E. coli DH5α Competent cells, and cultured to extract plasmid for preparing of DNA vaccines.
Recombinant proteins
Recombinant protein OmpA23–353 and Pal23–187 were expressed and purified similar to the protocol described previously [21]. In brief, the gene encoding the corresponding protein was cloned to the pGEX-6p-1 vector, the protein was expressed under the induction with 0.2 mM IPTG at 16 °C. GST-tagged proteins were harvested from cleared lysates with glutathione-Sepharose, and the GST tag was cleaved using PreScission Protease (GE Healthcare, Chicago, USA).The purified proteins were analyzed by SDS-PAGE and the concentration was determined by BCA assay.
Preparation of the CpG ODN 1826
Nuclease-resistant phosphorothioate-modified ODN 1826 (5′-TCCATGACGTTCCTGACGTT-3′) was synthesized by Invitrogen (Shanghai, China). The CpG ODN 1826 (5′-TCCAT-GACGTTCCTGACGTT-3′) were synthesized by Sangong (Shanghai, China) with a nuclease-resistant phosphorothioate backbone [22].
Intramuscular immunization of mice and serum collection
C57BL/6 mice were divided into five groups randomly (ten mice per group). After anaesthesia with pentobarbital, the quadriceps of mice were injected with 100 μl of DNA vaccine containing 15 μg of CpG ODN 1826 and 100 μg of pVAX1-OmpA, pVAX1-Pal and pVAX1-OmpA-Pal, respectively. As for the control group, mice were injected with equivalent pVAX1 blank plasmid and PBS. Mice in each group were inoculated in the same way on day 1, 15 and 22. The serum samples were collected 7 days after the final vaccination for further analysis. Two extra groups (three mice per group) immunized with pVAX1-OmpA and pVAX1-Pal without adjuvant were used as control for serum samples collection.
Measurement of mice survival against A. baumannii challenge
One week after the last immunization, all vaccinated mice were challenged with 20 μl of LAC-4 at two different dose (1.2 × 107 or 3.0 × 107 colony-forming units) or clinical strains (1.0 × 108 colony-forming units) of A. baumannii by intratracheal injection, survival rate from each group of mice was monitored for 7 days. Each mouse was observed and assessed through clinical symptom score [23] every 12 h. All dead mice were recorded.
Bacteria burden
Seven days after the last immunization, all vaccinated mice were challenged with 20 μl of LAC-4 at a sub-lethal dose (6 × 106 CFU) by intratracheal injection. 24 h after the infection, mice (ten mice/group) were sacrificed as described previously and lungs were taken out, weighed and homogenized in 2 ml PBS under sterile conditions. Serial 10-fold dilutions of lung homogenates were coated on Tryptic Soy Agar, and incubated at 37 °C for 16 h. All calculable CFUs were counted in a double-blinded manner.
Histological analysis
Lung tissue from the pneumonia model mice was removed under sterile conditions 24 h after infection. All lung samples were fixed with 10% formalin, embedded in paraffin, sliced, stained with hematoxylin and eosin (HE). Slices were observed at × 200 magnifications by the pathologist. All lung slices were quantitative analyzed through a score of 0–4 (no abnormality to most severe) according to the standard of exudate, edema, hemorrhage and inflammatory cell infiltration [24].
Evaluation of inflammation
Bronchoalveoar lavage fluid (BALF) was collected from infected mice 24 h after the challenge. In order to quantify neutrophil infiltration, BALF cells were stained with the following antibodies: PE/Cy7 anti-mouse CD45 antibody, PerCP-CY5.5 anti-mouse CD11b antibody and FITC anti-mouse Gr-1 antibody (Biolegend, Inc, San Diego, USA). Samples were analyzed by BD FACS Array software™ on BD flow cytometry (BD Biosciences, New Jersey, USA) to analyze the neutrophil infiltration. Then the supernatant of BALF was collected and used for cytokines detection. The levels of TNF-α in BALF supernatants were measured by using ELISA kit (Dakewe Biotech Co, Shenzhen, China), according to the manufacturer’s instructions.
Cytokine responses assay
Seven days after the last immunization, the mice were sacrificed by cervical dislocation under anesthetic with pentobarbital and their spleens were taken out under sterile conditions. Single-cell suspension of spleen was prepared according to standard procedure. For proliferative assay, spleen cells grown in a 96-well plate at a concentration of 4 × 105 living cells/200 μl per well. We added 30 μM/well of 5-bromo-2-deoxyuridine (BrdU, Roche, Germany) for measurement of lymphocyte proliferation. 10 mM phytohemagglutinin-A (PHA) (Gibco, USA) and PBS was served as positive and negative controls, recombinant protein OmpA and Pal, were added to the plates (10 μg/ml). Spleen cells were incubated at 37.0 °C with 5% CO2 for 96 h. Dyed incorporation was determined by measuring absorbance at 450 nm [25]. The stimulation index (SI) was calculated by dividing the absorbance of the stimulating cells by the absorbance of the unstimulating cells (SI = OD 450 of stimulated cells/OD 450 of unstimulated cells). Then the supernatant of spleen cells was collected and used for cytokines detection. The levels of INF-γ, IL-4 and IL-17A in supernatant were measured by using ELISA kit (Dakewe Biotech Co, China).
Statistical analysis
The data was expressed as mean ± Standard Deviation (SD) or mean ± Standard Error of Mean (SEM). The scores were marked blindly. The survival data was analyzed through Kaplan–Meier survival curves. To calculate P values, nonparametric Mann–Whitney test, log-rank test, Student’s t test, one-way ANOVA with Bonferroni correction were used depending on sample distribution and variation as mentioned in figure legends (SPSS statistics 18.0 and GraphPad Prism 6.0). P < 0.05 was considered as significant difference.
Results
Preparation of recombinant plasmids
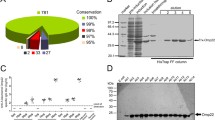
Three recombinant plasmids that encoding OmpA, Pal and the fusion protein OmpA-Pal were constructed in this study (Fig. 1a). The signal peptide of OmpA and Pal (both corresponding to residues 1–22) was predicted by SignalP 5.0 and was not included in these constructs. The fusion gene of OmpA-Pal was synthesized by Sangon, a flexible linker (GGGGS) was inserted between OmpA and Pal to avoid the possible interaction between the two proteins, the Kozak sequence (GCCACCATGG) was placed at the 5′ terminal of each fragment to ensure efficient translation of the resulting protein. Eukaryotic expression vector pVAX1 was used in this study because it was specifically designed for use in the development of DNA vaccines and was approved by FDA (Fig. 1b). PCR amplification combined with restriction enzyme digestion (BamH 1 and Xho 1) were performed to test the correctness of insertion. As shown in Fig. 3c, the sequences encoding OmpA, Pal and OmpA-Pal were approximate 1000 bp, 500 bp and 1500 bp in length on agarose gel when characterized by restriction endonuclease digestion. The inserts were sequenced and determined to be in complete agreement with the expected sequences, suggesting that all the recombinant plasmids were constructed correctly.
Construction and identification of recombinant plasmids. a Schematic presentation of DNA vaccines used in this study, three recombinant plasmids, termed pVAX1-OmpA, pVAX1-Pal and pVAX1-OmpA-Pal, were constructed to be used as DNA vaccines. b The construction map of recombinant plasmids, the Kozak sequence was added to the 5′ terminal of each insertion and then placed between the site of restrictive enzymes BamHI and XhoI of pVAX1 vector. c Identification of recombinant plasmids by agarose electrophoresis. Lane 1–3 represent recombinant plasmids pVAX1-OmpA, pVAX1-Pal and pVAX1-OmpA-Pal, respectively. Lane 4–6 represent the same plasmids digested by BamHI and XhoI
DNA vaccines improves survival rate in murine pneumonia model
To validate the protective efficacy of DNA vaccines, mice were immunized with pVAX1-OmpA, pVAX1-Pal and pVAX1-OmpA-Pal and CpG was used as adjuvant. As shown in Fig. 2a, compared with pVAX1 and PBS control group, mice vaccinated with DNA vaccines all exhibited higher survival rates at the end of the observation period. Moreover, when mice were challenged with 2 × LD50 of LAC-4 (1.2 × 107 CFUs/mouse), higher survival rates, but not statistical difference, were observed in pVAX1-OmpA-Pal immunized group when compared with those immunized with pVAX1-OmpA or pVAX1-Pal alone. When the challenging dose reached to 5 × LD50 (3.0 × 107 CFUs/mouse), mice immunized with pVAX1-OmpA-Pal showed a significantly higher survival rate than those immunized with pVAX1-OmpA or pVAX1-Pal alone (POmpA-Pal vs. OmpA = 0.0358 and POmpA-Pal vs. Pal = 0.0143, Fig. 2b). Furthermore, the survival rates in the pVAX1-OmpA-Pal group varies between 50 and 80%, and the death was delayed in mice immunized with all the DNA vaccines. These results clearly confirmed that OmpA and Pal could be used as promising candidate antigens for DNA vaccine development against A. baumannii infection, and immunization with pVAX1-OmpA-Pal resulted in improved protective efficacy when compared with pVAX1-OmpA or pVAX1-Pal immunized alone.
DNA vaccines improves survival rate in murine pneumonia model. C57BL/6 mice (n = 10) were immunized with DNA vaccines (pVAX1-OmpA, pVAX1-Pal and pVAX1-OmpA-Pal) plus CpG adjuvant and challenged with LAC-4 at 2 × LD50 (1.2 × 107 CFU/mouse, a) or 5 × LD50 (3.0 × 107 CFU/mouse, b) by intratracheal injection. The survival rate was monitored for 1 week. The P-value was calculated using the Mantel-Cox log-rank test
DNA vaccines protects mice from pneumonia by reducing bacterial burden and inflammation
Seven days after the last boost immunization, mice from all groups were challenged with a sub-lethal dose (6.0 × 106 CFU/mouse) of A. baumannii via the trachea to investigate the possible mechanism for DNA vaccines mediated protection. Firstly, the lungs from each group of mice were collected 24 h after challenge, and viable bacterial numbers were counted. As shown in Fig. 3a, the lungs from DNA vaccine groups showed significantly lower bacterial burden than the pVAX1 and PBS control group (P < 0.0001). Further, the bacterial burden was significantly reduced in the pVAX1-OmpA-Pal group compared with pVAX1-OmpA and pVAX1-Pal group (POmpA-Pal vs. OmpA = 0.0030 and POmpA-Pal vs. Pal = 0.0002, Fig. 3a). These results indicated that immunization with these DNA vaccines could partially reduce the colonization of A. baumannii in the lung of mice.
DNA vaccines protects mice from pneumonia by reducing bacterial burden and inflammation. a–f C57BL/6 mice were immunized with DNA vaccines (pVAX1-OmpA, pVAX1-Pal and pVAX1-OmpA-Pal) plus CpG adjuvant and infected with LAC-4 at 6.0 × 106 CFU/mouse by intratracheal injection. a The number of viable bacteria in the lungs of mice (n = 10) 24 h post-infection are shown. b Hematoxylin–eosin staining of lungs from immunized mice and control mice 24 h after infection, representative histopathological sections from 10 mice per group are shown (magnification = × 200). c Semi-quantification of lung inflammation in infected mice. Severity scores of lungs (n = 10) from immunized mice and control mice 24 h post-infection are shown. The data are presented as scatter plots. d Quantitative detection of proinflammatory cytokines (TNF-α) in infected mice (n = 10). (e–f) Evaluation of neutrophil infiltration in infected mice (n = 10). The bar represents the percentage (e) and the number (f) of neutrophils in the BALF of immunized mice 24 h post-challenge. The data (a, c–f) are shown as the mean ± SD. Multiple comparisons among different groups were calculated using one-way ANOVA (ns no significance)
We then evaluated the histological changes in mice, as shown in Fig. 3b, the lung slices of the DNA vaccine groups showed less extensive of lung lesions, alveolar edema, lymphocyte infiltration and structural damage caused by intensive inflammatory response compared with the pVAX1 and PBS control groups. Moreover, in the three DNA vaccine groups, there were more mononuclear cell infiltration, normal structure and clearer alveoli than the pVAX1 and PBS group. The results of severity score showed that the severity of lung injury in DNA vaccine groups was significantly lower than that in pVAX1 and PBS control group (Fig. 3c), suggesting that mice immunized with DNA vaccines showed decreased inflammatory response in lungs.
In the BALF, inflammatory biomarkers including pro-inflammatory cytokine TNF-α, neutrophil percentages and absolute values were measured 24 h after the pulmonary infection. As shown in Fig. 3d, the level of TNF-α in mice immunized with DNA vaccine was significantly lower than that of mice immunized with pVAX1 or PBS, and the level of TNF-α from mice immunized with pVAX1-OmpA-Pal was significantly lower than that of the pVAX1-Pal (P = 0.0078, Fig. 3d). Meanwhile, the level of neutrophil percentages and absolute values about neutrophil in the BALF of mice immunized with DNA vaccine was significantly lower than that of mice immunized with PBS or pVAX1 (Fig. 3e, f). In addition, mice immunized with pVAX1-OmpA-Pal cause a decrease in neutrophil percentages and absolute values compared with mice immunized with pVAX1-Pal (Pneutrophil percentages = 0.0003, Pneutrophil absolute values = 0.0233). These results were consistent with reduced cell infiltration observed by histological analysis.
About all, the immune protection of DNA vaccines was correlated with reduced viable bacterial numbers, pathological change, pro-inflammatory cytokine production and neutrophil infiltration in the lung tissue in mice.
DNA vaccines elicits a mixed Th1, Th2 and Th17 responses
In order to clarify the protective efficacy of DNA vaccine, corresponding recombinant proteins were used as antigens to stimulate the spleen cells of immunized mice, the proliferation reaction and cytokine level were measured in vitro. Firstly, the stimulation index (SI) of spleen cells from immunized mice was measured. As shown in Fig. 4a, the SI of the spleen cells from mice immunized with pVAX1-OmpA, pVAX1-Pal or pVAX1-OmpA -Pal was significantly higher than that of the two control group (P < 0.0001), and the SI of the spleen cells from mice immunized with pVAX1-OmpA-Pal was significantly higher than that of the pVAX1-Pal (P = 0.0077). However, there was no difference between mice immunized with pVAX1-OmpA-Pal and pVAX1-OmpA (Fig. 4a).
Analysis of spleen cells proliferation and cytokine secretion. a Lympho-proliferative activity of mouse spleen cells (n = 5) after in vitro stimulation with corresponding recombinant protein (OmpA or/and Pal) for 72 h, respectively. Proliferation was measured using the bromodeoxyuridine (BrdU) labeling method. The stimulation index (SI) was calculated by dividing the absorbance of the stimulating cells by the absorbance of the unstimulating cells. b–d Comparison of cytokine production by antigens stimulated spleen cells from immunized and control mice. Two weeks after the final immunization, spleens (n = 5) were processed and stimulated with corresponding recombinant protein, and the levels of IL-4 (b), IFN-γ (c), and IL-17 (d) in each culture supernatant were measured after 72 h. Differences were compared to determine their statistical significance using one-way ANOVA followed by Tukey’s multiple comparison test (ns no significance)
Then, the level of cytokines secreted by stimulated spleen cells were measured by ELISA. The spleen cells from the DNA vaccine immunized mice induced significantly higher level of IL4, IFN-γ and IL-17A when stimulated with the corresponding recombinant proteins than did the pVAX1 and PBS control group (P < 0.0001, Fig. 4b–d). Moreover, the spleen cells from mice immunized with pVAX1-OmpA-Pal induced the highest level of IFN-γ after corresponding recombinant proteins stimulation (P < 0.0001, Fig. 4b–d). Besides, the level of IL-4 and IL-17A of the spleen cells from mice immunized with pVAX1-OmpA-Pal was significantly higher than that of the pVAX1-Pal (PIL-4 = 0.0004, PIL-17A = 0.0004). In contrast, there was no statistical difference about the level of IL-4 and IL-17A between pVAX1-OmpA-Pal and pVAX1-OmpA group. These results demonstrated that immunize with DNA vaccines was efficient to induce cellular response.
DNA vaccines induce high level of humoral response
In order to determine the immunogenicity of DNA vaccines, serum antibody levels against both OmpA and Pal antigens were evaluated after the final immunization. As shown in Fig. 5a, c the level of antigen specific antibody (IgG) in all DNA vaccine groups were dramatically higher than in the pVAX1 and PBS group (P < 0.0001). However, there was no difference between pVAX1-OmpA-Pal and single DNA vaccine groups. Moreover, CpG adjuvant was efficient to enhance the humoral response induced by DNA vaccines, as the antigen specific antibody was significantly higher in the serum of mice immunized with DNA vaccines plus adjuvant compared to DNA vaccines alone (Fig. 5a, c).
Analysis of antibody responses. (a–d) C57BL/6 mice were immunized with DNA vaccines (pVAX1-OmpA, pVAX1-Pal and pVAX1-OmpA-Pal) with (n = 10) or without (n = 3) CpG adjuvant, PBS and pVAX1 were used as negative control (n = 10). Serum was collected from mice on day 28 and diluted by PBS at a ratio of 1:1000. a Comparison of serum total IgG against OmpA. b Comparison of serum IgG1 and IgG2a subtypes against OmpA. c Comparison of serum total IgG against Pal. d Comparison of serum IgG1 and IgG2a subtypes against Pal. Data were shown as the mean ± SD. Multiple comparisons among different groups were analyzed using one-way ANOVA (ns no significance)
Furthermore, to explore the type of specific antibody response induced by each antigen, the level of IgG subtypes (IgG1 and IgG2a) in serum was evaluated by ELISA. When mice were immunized with pVAX1-OmpA alone, the level of IgG1 was significantly higher than IgG2a, indicating a Th2 biased response was induced. When CpG was used as adjuvant, a relatively balanced Th1/Th2 response was observed, this is in consistence with the ability of CpG to induce cellular response (Fig. 5b). In contrast, mice immunized with pVAX1-Pal alone showed higher level of IgG2a than pVAX1-OmpA, and a balanced Th1/Th2 response was induced without adjuvant, which was also observed in a previous report [26], suggesting that Pal is efficient to induce cellular response (Fig. 5d). Further, when CpG was used as adjuvant, there is no difference in the level of IgG1 in all groups, but the level of IgG2a was significantly increased, suggesting that the increased level of humoral response induced by CpG was mainly attributed to increased level of IgG2a. Moreover, when adjuvant was used, there is no difference in the level of IgG1 and IgG2a in mice immunized with pVAX1-OmpA-pal compared with those immunized with pVAX1-OmpA or pVAX1-Pal alone (Fig. 5b, d), which was similar to the result of total IgG. These results indicate that the protective efficacy of DNA vaccines was not mediated by humoral response alone, other mechanisms, such as cellular and innate immune response, also participated in the protective efficacy.
DNA vaccine immunization provide cross-protective efficacy in mice challenged with clinical strains of A. baumannii
There is report that the sequences of both OmpA and Pal were highly conserved among different species and different strains of A. baumannii [27, 28], and we have aligned the sequence of the two antigens from different strains of A. baumannii, as shown in supplementary Figs. 1 and 2, the sequence of the two antigens almost completely conserved among different strains. Thus we wonder if DNA vaccine immunization could provide protective effect against challenge with different clinical strains of A. baumannii. In this study, three clinical strains were chosen to further test the protective efficacy of the DNA vaccine. As shown in Fig. 6, mice were immunized with DNA vaccine and challenged with 1.0 × 108 CFUs of different clinical strains of A. baumannii. One week after infection, all the three clinical strains exhibited different pathogenicity and virulence in mice (SJZ18 < SJZ04 < SJZ28), as indicated by the survival rates in the PBS control group. DNA vaccine immunization significantly decreased the death in DNA vaccine immunized mice as compared with control group. This result strongly indicated that this DNA vaccine yielded broad protective efficacy after immunization. Given the high conservation of the two antigens, the different efficacy observed between LAC-4 and the clinical strains may due to the potential masking of surface antigens by the capsule of A. baumannii, which is crucial in development of a generalizable vaccine or therapeutic antibody [29].
Immunization with DNA vaccine provide cross protective efficacy in mice pneumonia model. C57BL/6 mice (n = 10) were immunized with pVAX1-OmpA-Pal plus CpG adjuvant and challenged with different clinical strains of A. baumannii at 1.0 × 108 CFU/mouse by intratracheal injection. The survival rate was monitored for 1 week. The P-value was calculated using the Mantel-Cox log-rank test
Discussion
The OmpA family proteins are abundant and highly conserved among different species of Gram-negative bacteria, and they have been reported to be essential in bacterial growth and infection. For example, they are involved in bacterial virulence, adhesion, invasion and serve as adaptor proteins to interact with surface receptors on host cells [27, 30]. More importantly, these proteins exhibit high immunogenicity and OmpA specific antibodies are reported to be protective in animal experiments [15, 31]. These properties make them promising candidates for vaccine development, and OmpA family proteins from a variety of species have been reported to be able to induce protective immunity against bacterial infection, such as P. aeruginosa, S. flexneri, C. abortus, E. coli and K. pneumonia [31,32,33,34,35]. OmpA from A. baumannii was also reported to be able to induce high titers of anti-OmpA antibodies and markedly improved survival and reduced tissue bacterial burden in mice infected intravenously [14, 15]. We have cloned the gene encoding OmpA of A. baumannii to a number of expression plasmids, but it was difficult to get soluble fractions of this protein, mainly because it is a membrane associated protein. Finally, we cloned it to the pGEX6p-1 vector and expressed it as a GST-OmpA fusion protein, most of OmpA was precipitated when the GST tag was digested, only a little fraction was soluble in solution, thus it is difficult to be used as ideal candidate antigens for further development of vaccines. Similar to OmpA, expression of the full length of Pal was also not successful. So in this study, we aimed to construct a DNA vaccine based on the sequences of the two proteins.
Adjuvants is a critical component for an effective vaccine. In this study, CpG was selected as adjuvant to enhance the immune response of DNA vaccine against A. baumannii infection. For one thing, CpG is the most commonly used adjuvant for DNA vaccine development [36, 37]. For another, there is report that neutrophil mediated host defenses induced by Th1-type cytokines may be essential for the elimination of A. baumannii [38], and CpG was capable of stimulating the secretion of cytokines such as IFN-γ and IL-12, both of which support the generation of Th1-dependent immune responses. Our results clearly showed that immune with DNA vaccine with CpG resulted in higher level of antigen specific IgG2a, which is a marker of a Th1 immune response, and the level of IFN-γ was increased followed by DNA vaccines immunization, which also confirmed that a Th1 immune response was induced. Besides, CpG was also reported to play an important role in antibody production and was able to improve mucosal immune responses [39].
It has been reported that immunization of mice with recombinant OmpA formulated with aluminum hydroxide adjuvant markedly improved survival and reduced tissue bacterial burden, and the levels of antigen specific antibody correlated with survival rate in mice [40], indicating that OmpA specific antibody is essential for the protective efficacy. Another study reported that OmpA from A. baumannii induces differentiation of CD4+ T cells toward a Th1 polarizing phenotype through the activation of dendritic cells [41]. In this study, we found that immune of OmpA without adjuvant was able to induce high level of humoral response, and a Th2-bialsd response was induced as indicted by antibody subtype assay. When CpG was used as adjuvant, elevated antibody level was observed and a Th1 immune response was enhanced. The difference observed in these studies may be attributed to the use of different adjuvants.
Previous studies observed that intravenously immunization with recombinant Pal was able to elicit a strong humoral response due to the activation of Th2 lymphocytes, but it does not evoke an efficient cellular response. In contrast, intramuscular immunization of a DNA vaccine based on pal was found to induce a Th1-biased response [26]. A later study reported that passive immunization with E. coli Pal specific antibody did not protect mice in a mice sepsis model [42], indicating Pal mediated protection is not mediated by humoral response. In this study, in order to induce a high level of Th1 response, we developed DNA vaccine based on Pal and used CpG as adjuvant, the results showed that high level of Pal specific antibodies was detected upon immunization with Pal, which was consistent with results reported previously [26]. Meanwhile, immunization with Pal alone mainly induce a Th2 response, a relatively lower level of Th1 response was also observed. When CpG was formulated with Pal, a balanced immune response was elicited, which may be essential for Pal mediated protection.
In this study, we have add the Kozak sequence to the 5′ terminal of the plasmid to enhance the expression of the antigens. However, we have no OmpA or Pal specific antibody for the moment, and add any other tags, such as GFP and HA, to the N- or C-terminal of the antigens may change the conformation of the resulting protein, thus the level of expression of OmpA and Pal was not determined following vaccination, which is a limitation of this study. When testing the protective efficacy, plasmid encoding the two antigens was significantly higher than the empty plasmid, indicating that both of OmpA and Pal were expressed, but further quantification of the level of expression is required.
Although two antigens were used in this study, and a mixed humoral and cellular response was induced upon immunization, only 50–80% mice were survived after lethal challenge with A. baumannii. To improve the protective efficacy, further studies will focused on identify other candidate antigens for vaccine development. Meanwhile, we will try to change the delivery system of DNA vaccines, such as liposomes, microspheres, nanoparticles, dendrimers or micellar systems, which has reported to be effective to enhance the efficacy of vaccines [43]. Finally, we will also try to compare the efficacy of the vaccine by using different administration route, such as intranasal vaccination, which was confirmed to induce higher IgA and cellular response to a DNA vaccine and improves protection against pulmonary mycobacterial challenge [44].
References
Howard A, O’Donoghue M, Feeney A, Sleator RD (2012) Acinetobacter baumannii: an emerging opportunistic pathogen. Virulence 3:243–250
Falagas ME, Bliziotis IA, Siempos II (2006) Attributable mortality of Acinetobacter baumannii infections in critically ill patients: a systematic review of matched cohort and case-control studies. Crit Care 10:R48
Papadimitriou-Olivgeris M, Fligou F, Spiliopoulou A, Koutsileou K, Kolonitsiou F et al (2017) Risk factors and predictors of carbapenem-resistant Pseudomonas aeruginosa and Acinetobacter baumannii mortality in critically ill bacteraemic patients over a 6-year period (2010–2015): antibiotics do matter. J Med Microbiol 66:1092–1101
Santajit S, Indrawattana N (2016) Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res Int 2016:2475067
Lin MF, Lan CY (2014) Antimicrobial resistance in Acinetobacter baumannii: from bench to bedside. World J Clin Cases 2:787–814
Moriel DG, Beatson SA, Wurpel DJ, Lipman J, Nimmo GR et al (2013) Identification of novel vaccine candidates against multidrug-resistant Acinetobacter baumannii. PLoS ONE 8:e77631
Pachon J, McConnell MJ (2014) Considerations for the development of a prophylactic vaccine for Acinetobacter baumannii. Vaccine 32:2534–2536
Shahid F, Ashraf ST, Ali A (2019) Reverse vaccinology approach to potential vaccine candidates against Acinetobacter baumannii. Methods Mol Biol 1946:329–336
Chen W (2015) Current advances and challenges in the development of Acinetobacter vaccines. Hum Vaccin Immunother 11:2495–2500
Chiang MH, Sung WC, Lien SP, Chen YZ, Lo AF et al (2015) Identification of novel vaccine candidates against Acinetobacter baumannii using reverse vaccinology. Hum Vaccin Immunother 11:1065–1073
Lee IM, Yang FL, Chen TL, Liao KS, Ren CT et al (2018) Pseudaminic Acid on exopolysaccharide of Acinetobacter baumannii plays a critical role in phage-assisted preparation of glycoconjugate vaccine with high antigenicity. J Am Chem Soc 140:8639–8643
McConnell MJ, Rumbo C, Bou G, Pachon J (2011) Outer membrane vesicles as an acellular vaccine against Acinetobacter baumannii. Vaccine 29:5705–5710
Lu S, Wang S, Grimes-Serrano JM (2008) Current progress of DNA vaccine studies in humans. Expert Rev Vaccin 7:175–191
Lin L, Tan B, Pantapalangkoor P, Ho T, Hujer AM et al (2013) Acinetobacter baumannii rOmpA vaccine dose alters immune polarization and immunodominant epitopes. Vaccine 31:313–318
Zhang X, Yang T, Cao J, Sun J, Dai W et al (2016) Mucosal immunization with purified OmpA elicited protective immunity against infections caused by multidrug-resistant Acinetobacter baumannii. Microb Pathog 96:20–25
Yeo KJ, Lee WC, Lee S, Hwang E, Park JS et al (2017) d-Stereoisomer preference of the OmpA-like domain of Pal in peptidoglycan of Acinetobacter baumannii. Process Biochem 55:110–115
Godlewska R, Wisniewska K, Pietras Z, Jagusztyn-Krynicka EK (2009) Peptidoglycan-associated lipoprotein (Pal) of Gram-negative bacteria: function, structure, role in pathogenesis and potential application in immunoprophylaxis. FEMS Microbiol Lett 298:1–11
Cordwell SJ, Len AC, Touma RG, Scott NE, Falconer L et al (2008) Identification of membrane-associated proteins from Campylobacter jejuni strains using complementary proteomics technologies. Proteomics 8:122–139
Murphy TF, Kirkham C, Lesse AJ (2006) Construction of a mutant and characterization of the role of the vaccine antigen P6 in outer membrane integrity of nontypeable Haemophilus influenzae. Infect Immun 74:5169–5176
Harris G, Kuo Lee R, Lam CK, Kanzaki G, Patel GB et al (2013) A mouse model of Acinetobacter baumannii-associated pneumonia using a clinically isolated hypervirulent strain. Antimicrob Agents Chemother 57:3601–3613
Yang F, Gu J, Zou J, Lei L, Jing H et al (2018) PA0833 is an OmpA C-like protein that confers protection against Pseudomonas aeruginosa infection. Front Microbiol 9:1062
Yan W, Sun TY, Yang CM, Jia M, Li J et al (2015) CpG ODN 1826 enhances radiosensitivity of the human lung cancer cell line A549 in a rat model. Genet Mol Res 14:9804–9812
Moura-Alves P, Fae K, Houthuys E, Dorhoi A, Kreuchwig A et al (2014) AhR sensing of bacterial pigments regulates antibacterial defence. Nature 512:387–392
Zuo QF, Yang LY, Feng Q, Lu DS, Dong YD et al (2013) Evaluation of the protective immunity of a novel subunit fusion vaccine in a murine model of systemic MRSA infection. PLoS ONE 8:e81212
Behrouz B, Mahdavi M, Amirmozafari N, Fatemi MJ, Irajian G et al (2016) Immunogenicity of Pseudomonas aeruginosa recombinant b-type fagellin as a vaccine candidate: protective efficacy in a murine burn wound sepsis model. Burns. https://doi.org/10.1016/j.burns.2016.03.015
Yoon WS, Park SH, Park YK, Park SC, Sin JI et al (2002) Comparison of responses elicited by immunization with a Legionella species common lipoprotein delivered as naked DNA or recombinant protein. DNA Cell Biol 21:99–107
Confer AW, Ayalew S (2013) The OmpA family of proteins: roles in bacterial pathogenesis and immunity. Vet Microbiol 163:207–222
Liang MD, Bagchi A, Warren HS, Tehan MM, Trigilio JA et al (2005) Bacterial peptidoglycan-associated lipoprotein: a naturally occurring toll-like receptor 2 agonist that is shed into serum and has synergy with lipopolysaccharide. J Infect Dis 191:939–948
Wang-Lin SX, Olson R, Beanan JM, MacDonald U, Balthasar JP et al (2017) The capsular polysaccharide of Acinetobacter baumannii is an obstacle for therapeutic passive immunization strategies. Infect Immun 85:e005971
Smith SG, Mahon V, Lambert MA, Fagan RP (2007) A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol Lett 273:1–11
Pore D, Chakrabarti MK (2013) Outer membrane protein A (OmpA) from Shigella flexneri 2a: a promising subunit vaccine candidate. Vaccine 31:3644–3650
Jeannin P, Magistrelli G, Goetsch L, Haeuw JF, Thieblemont N et al (2002) Outer membrane protein A (OmpA): a new pathogen-associated molecular pattern that interacts with antigen presenting cells-impact on vaccine strategies. Vaccine 20(Suppl 4):A23–27
Ayalew S, Shrestha B, Montelongo M, Wilson AE, Confer AW (2011) Immunogenicity of Mannheimia haemolytica recombinant outer membrane proteins serotype 1-specific antigen, OmpA, OmpP2, and OmpD15. Clin Vaccin Immunol 18:2067–2074
Hounsome JD, Baillie S, Noofeli M, Riboldi-Tunnicliffe A, Burchmore RJ et al (2011) Outer membrane protein A of bovine and ovine isolates of Mannheimia haemolytica is surface exposed and contains host species-specific epitopes. Infect Immun 79:4332–4341
Tang X, Wang H, Liu F, Sheng X, Xing J et al (2017) Outer membrane protein A: an immunogenic protein induces highly protective efficacy against Vibrio ichthyoenteri. Microb Pathog 113:152–159
Qiu Z, Xing L, Zhang X, Qiang X, Xu Y et al (2017) CpG oligodeoxynucleotides augment antitumor efficacy of folate receptor alpha based DNA vaccine. Oncol Rep 37:3441–3448
Zhao F, Liu S, Zhang X, Yu J, Zeng T et al (2013) CpG adjuvant enhances the mucosal immunogenicity and efficacy of a Treponema pallidum DNA vaccine in rabbits. Hum Vaccin Immunother 9:753–760
Tsuchiya T, Nakao N, Yamamoto S, Hirai Y, Miyamoto K et al (2012) NK1.1(+) cells regulate neutrophil migration in mice with Acinetobacter baumannii pneumonia. Microbiol Immunol 56:107–116
Bode C, Zhao G, Steinhagen F, Kinjo T, Klinman DM (2011) CpG DNA as a vaccine adjuvant. Expert Rev Vaccin 10:499–511
Luo G, Lin L, Ibrahim AS, Baquir B, Pantapalangkoor P et al (2012) Active and passive immunization protects against lethal, extreme drug resistant-Acinetobacter baumannii infection. PLoS ONE 7:e29446
Lee JS, Lee JC, Lee CM, Jung ID, Jeong YI et al (2007) Outer membrane protein A of Acinetobacter baumannii induces differentiation of CD4+ T cells toward a Th1 polarizing phenotype through the activation of dendritic cells. Biochem Pharmacol 74:86–97
Valentine CH, Hellman J, Beasley-Topliffe LK, Bagchi A, Warren HS (2006) Passive immunization to outer membrane proteins MLP and PAL does not protect mice from sepsis. Mol Med 12:252–258
Saroja C, Lakshmi P, Bhaskaran S (2011) Recent trends in vaccine delivery systems: a review. Int J Pharm Investig 1:64–74
Wu M, Zhao H, Li M, Yue Y, Xiong S et al (2017) Intranasal vaccination with mannosylated chitosan formulated DNA vaccine enables robust IgA and cellular response induction in the lungs of mice and improves protection against pulmonary mycobacterial challenge. Front Cell Infect Microbiol 7:445
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 81771778).
Author information
Authors and Affiliations
Contributions
XW, JYZ and QZ designed the research; LL, FY and JTZ conducted the experiments, analyzed the data, wrote the main manuscript text and prepared the figures and tables, HJ, JZ and WX helped to conduct the experiments, JYZ, LL and XW contributed to writing the manuscript and supervised the project. FY and QZ helped with the discussion of results and manuscript refinement.
Corresponding authors
Ethics declarations
Conflict of interest
We declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lei, L., Yang, F., Zou, J. et al. DNA vaccine encoding OmpA and Pal from Acinetobacter baumannii efficiently protects mice against pulmonary infection. Mol Biol Rep 46, 5397–5408 (2019). https://doi.org/10.1007/s11033-019-04994-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-019-04994-2