Abstract
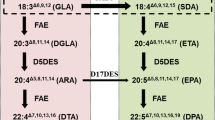
An important alternative source of fish oil is its production by plants through metabolic engineering. To produce eicosapentaenoic acid (EPA, 20:5n-3) in peanut through the alternative Δ8-pathway, a plant expression vector containing five heterologous genes driven by the constitutive 35S promoter respectively, namely, ∆9-elongase (Isochrysis galbana), ∆8-desaturase (Euglena gracilis), ∆5-desaturase (Mortierella alpina), ∆15-desaturase (Arabidopsis thaliana) and ∆17-desaturase (Phytophthora infestans) were transferred into peanut through Agrobacterium-mediated transformation method. The gas chromatography results indicated that the average content of EPA in the leaves of the transgenic lines was 0.68%, and the highest accumulation of EPA in an individual line reached 0.84%. This finding indicates that it is feasible to synthesize EPA in peanut through metabolic engineering and lays the foundations for the production of very-long-chain polyunsaturated fatty acids (VLCPUFAs) in peanut seeds.





Similar content being viewed by others
Abbreviations
- EPA:
-
Eicosapentaenoic acid
- VLCPUFAs:
-
Very-long-chain polyunsaturated fatty acids
- DHA:
-
Docosahexaenoic acid
- LA:
-
Linoleic acid
- ALA:
-
α-Linolenic acid
- 6-BA:
-
6-Benzyl aminopurine
- NAA:
-
1-Naphthylacetic acid
- MCS:
-
Multiple cloning site
- CTAB:
-
Cetyltrimethyl ammonium bromide
- Elo :
-
Elongase
- Des :
-
Desaturase
- GC:
-
Gas chromatography
- FAMEs:
-
Fatty acid methyl esters
- GC–MS:
-
Gas chromatograph–mass spectrometer
- ETrA:
-
Eicosatrienoic acid
References
Gill I, Valivety R (1997) Polyunsaturated fatty acids, part 1: occurrence, biological activities and applications. Trends Biotechnol 15:401–409
Lauritzen L, Hansen H, Jørgensen M, Michaelsen K (2001) The essentiality of long chain n-3 fatty acids in relation to development and function of the brain and retina. Prog Lipid Res 40:1–94
Thies F, Garry JM, Yaqoob P, Rerkasem K, Williams J, Shearman CP, Gallagher PJ, Calder PC, Grimble RF (2003) Association of n-3 polyunsaturated fatty acids with stability of atherosclerotic plaques: a randomised controlled trial. Lancet 361:477–485
Abeywardena P (2011) Role of ω3 long chain polyunsaturated fatty acids in reducing cardio-metabolic risk factors. Endocr Metab Immune Disord Drug Targets 11:232–246
Sprecher H (2000) Metabolism of highly unsaturated n-3 and n-6 fatty acids. Biochim Biophys Acta 1486:219–231
Pawlosky RJ, Hibbeln JR, Novotny JA, Salem N (2001) Physiological compartmental analysis of α-linolenic acid metabolism in adult humans. J Lipid Res 42:1257–1265
Snyder CL, Yurchenko OP, Siloto RM, Chen X, Liu Q, Mietkiewska E, Weselake RJ (2009) Acyltransferase action in the modification of seed oil biosynthesis. New Biotechnol 26:11–16
Yurchenko OP, Nykiforuk CL, Moloney MM, Ståhl U, Banaś A, Stymne S, Weselake RJ (2009) A 10-kDa acyl-CoA-binding protein (ACBP) from Brassica napus enhances acyl exchange between acyl-CoA and phosphatidylcholine. Plant Biotechnol J 7:602–610
Abbadi A, Domergue F, Bauer J, Napier J, Welti R, Zähringer U, Cirpus P, Heinz E (2004) Biosynthesis of very-long-chain polyunsaturated fatty acids in transgenic oilseeds: constraints on their accumulation. Plant Cell Online 16:2734–2748
Kinney A, Cahoon E, Damude H, Hitz W, Liu ZB, Kolar C (2004) Production of very long chain polyunsaturated fatty acids in oilseed plants, Google Patents 2004
Kinney A, Cahoon E, Damude H, Hitz W, Liu ZB, Kolar C (2011) Production of very long chain polyunsaturated fatty acids in oil seed plants, Google Patents 2011
Wu G, Truksa M, Datla N, Vrinten P, Bauer J, Zank T, Cirpus P, Heinz E, Qiu X (2005) Stepwise engineering to produce high yields of very long-chain polyunsaturated fatty acids in plants. Nat Biotechnol 23:1013–1017
Cheng B, Wu G, Vrinten P, Falk K, Bauer J, Qiu X (2010) Towards the production of high levels of eicosapentaenoic acid in transgenic plants: the effects of different host species, genes and promoters. Transgenic Res 19:221–229
Petrie JR, Shrestha P, Mansour MP, Nichols PD, Liu Q, Singh SP (2010) Metabolic engineering of omega-3 long-chain polyunsaturated fatty acids in plants using an acyl-CoA ∆6-desaturase with ω3-preference from the marine microalga Micromonas pusilla. Metab Eng 12:233–240
Petrie JR, Shrestha P, Zhou XR, Mansour MP, Liu Q, Belide S, Nichols PD, Singh SP (2012) Metabolic engineering plant seeds with fish oil-like levels of DHA. PLoS ONE 7:e49165
Ruiz-Lopez N, Haslam RP, Napier JA, Sayanova O (2014) Successful high-level accumulation of fish oil omega-3 long-chain polyunsaturated fatty acids in a transgenic oilseed crop. Plant J 77:198–208
Qi B, Fraser T, Mugford S, Dobson G, Sayanova O, Butler J, Napier JA, Stobart AK, Lazarus CM (2004) Production of very long chain polyunsaturated omega-3 and omega-6 fatty acids in plants. Nat Biotechnol 22:739–745
Liu J, Ma Y, Sun Q, Wu X, Li X, Sun M, Li Y, Li X, Qi B (2014) Production of very long chain polyunsaturated fatty acids in cotton. Acta Agron Sin 40:86–92
Ruiz-Lopez N, Haslam RP, Usher S, Napier JA, Sayanova O (2015) An alternative pathway for the effective production of the omega-3 long-chain polyunsaturates EPA and ETA in transgenic oilseeds. Plant Biotechnol J 13:1264–1275
Özcan M, Seven S (2003) Physıcal and chemıcal analysıs and fatty acıd composıtıon of peanut, peanut oıl and peanut butter from ÇOM and NC-7 cultıvars. Grasas Aceites 54:12–18
Sharma KK, Anjaiah V (2000) An efficient method for the production of transgenic plants of peanut (Arachis hypogaea L.) through Agrobacterium tumefaciens-mediated genetic transformation. Plant Sci 159:7–19
Klein TM, Wolf E, Wu R, Sanford J (1987) High-velocity microprojectiles for delivering nucleic acids into living cells. Nature 327:70–73
Rohini V, Rao KS (2001) Transformation of peanut (Arachis hypogaea L.) with tobacco chitinase gene: variable response of transformants to leaf spot disease. Plant Sci 160:889–898
Bhatnagar-Mathur P, Devi MJ, Reddy DS, Lavanya M, Vadez V, Serraj R, Yamaguchi-Shinozaki K, Sharma KK (2007) Stress-inducible expression of At DREB1A in transgenic peanut (Arachis hypogaea L.) increases transpiration efficiency under water-limiting conditions. Plant Cell Rep 26:2071–2082
Tiwari S, Mishra DK, Singh A, Singh P, Tuli R (2008) Expression of a synthetic crylEC gene for resistance against Spodoptera litura in transgenic peanut (Arachis hypogaea L.). Plant Cell Rep 27:1017–1025
Yang CY, Chen SY, Duan GC (2011) Transgenic peanut (Arachis hypogaea L.) expressing the urease subunit B gene of Helicobacter pylori. Curr Microbiol 63:387–391
Sun Q, Liu J, Li Y, Zhang Q, Shan S, Li X, Qi B (2013) Creation and validation of a widely applicable multiple gene transfer vector system for stable transformation in plant. Plant Mol Biol 83:391–404
Ye X, Al-Babili S, Klöti A, Zhang J, Lucca P, Beyer P, Potrykus I (2000) Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287:303–305
Liu X, Yang W, Mu B, Li S, Li Y, Zhou X, Zhang C, Fan Y, Chen R (2018) Engineering of ‘Purple Embryo Maize’ with a multigene expression system derived from a bidirectional promoter and self-cleaving 2A peptides. Plant Biotechnol J 16:1107–1109
Zhu Q, Yu S, Zeng D, Liu H, Wang H, Yang Z, Xie X, Shen R, Tan J, Li H, Zhao X, Zhang Q, Chen Y, Guo J, Chen L, Liu YG (2017) Development of ‘‘Purple Endosperm Rice’’ by engineering anthocyanin biosynthesis in the endosperm with a high-efficiency transgene stacking system. Mol Plant 10:918–929
Pereira SL, Leonard AE, Mukerji P (2003) Recent advances in the study of fatty acid desaturases from animals and lower eukaryotes. Prostaglandins Leukot Essent Fatty Acids 68:97–106
Sun Q, Liu J, Zhang Q, Qing X, Dobson G, Li X, Qi B (2013) Characterization of three novel desaturases involved in the delta-6 desaturation pathways for polyunsaturated fatty acid biosynthesis from Phytophthora infestans. Appl Microbiol Biotechnol 97:7689–7697
Haslam RP, Ruiz-Lopez N, Eastmond P, Moloney M, Sayanova O, Napier JA (2013) The modification of plant oil composition via metabolic engineering-better nutrition by design. Plant Biotechnol J 11:157–168
Chen Y, Meesapyodsuk D, Qiu X (2014) Transgenic production of omega-3 very long chain polyunsaturated fatty acids in plants: accomplishment and challenge. Biocatal Agric Biotechnol 3:38–43
Petrie JR, Shrestha P, Belide S, Kennedy Y, Lester G, Liu Q, Divi UK, Mulder RJ, Mansour MP, Nichols PD (2014) Metabolic engineering Camelina sativa with fish oil-like levels of DHA. PLoS ONE 9:e85061
Xia F, Li X, Li X, Zheng D, Sun Q, Liu J, Li Y, Hua J, Qi B (2016) Elevation of the yields of very long chain polyunsaturated fatty acids via minimal codon optimization of two key biosynthetic enzymes. PLoS ONE 11:e0158103
Lu C, Napier JA, Clemente TE, Cahoon EB (2011) New frontiers in oilseed biotechnology: meeting the global demand for vegetable oils for food, feed, biofuel, and industrial applications. Curr Opin Biotechnol 22:252–259
Acknowledgements
This study was funded by the Shandong Province Peanut Seed Industry Project, the National Natural Science Foundation of China (31601336, 31271757) and the National Opening Project of State Key Laboratory of Crop Biology (2016KF09). The sequencing and assembly were performed by the Beijing Genomics Institute (BGI).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, C., Qing, X., Yu, M. et al. Production of eicosapentaenoic acid (EPA, 20:5n-3) in transgenic peanut (Arachis hypogaea L.) through the alternative Δ8-desaturase pathway. Mol Biol Rep 46, 333–342 (2019). https://doi.org/10.1007/s11033-018-4476-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-018-4476-1