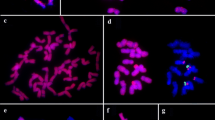
Abstract
Repetitive DNA sequences have been widely used in cytogenetic analyses. The use of gene sequences with a low-copy-number, however, is little explored especially in plants. To date, the karyotype details in Brachiaria spp. are limited to the location of rDNA sites. The challenge lies in developing new probes based on incomplete sequencing data for the genus or complete sequencing of related species, since there are no model species with a sequenced genome in Brachiaria spp. The present study aimed at the physical location of conserved genes in chromosomes of Brachiaria ruziziensis, Brachiaria brizantha, and Brachiaria decumbens using RNAseq data, as well as sequences of Setaria italica and Sorghum bicolor through the fluorescent in situ hybridization technique. Five out of approximately 90 selected sequences generated clusters in the chromosomes of the species of Brachiaria studied. We identified genes in synteny with 5S and 45S rDNA sites, which contributed to the identification of chromosome pairs carrying these genes. In some cases, the species of Brachiaria evaluated had syntenic segments conserved across the chromosomes. The use of genomic sequencing data is essential for the enhancement of cytogenetic analyses.
Similar content being viewed by others
References
Wang CJ, Harper L, Cande WZ (2006) High-resolution single-copy gene fluorescence in situ hybridization and its use in the construction of a cytogenetic map of maize chromosome 9. Plant Cell 18:529–544. https://doi.org/10.1105/tpc.105.037838
Huang PL, Hahlbrock K, Somssich IE (1988) Detection of a single-copy gene on plant chromosomes by in situ hybridization. Mol Genet Genom 211:143 147. https://doi.org/10.1007/BF00338405
Dong H, Quick JS (1995) Detection of 2.6 kb single-low copy DNA sequence on chromosomes of wheat (Triticum aestivum) and rye (Secale cereale). Genome 38:246–249. https://doi.org/10.1139/g95-030
Fransz PF et al (1996) Detection of single-copy genes and chromosome rearrangements in Petunia hybrida by fluorescence in situ hybridization. Plant J 9:767–774. https://doi.org/10.1046/j.1365-313X.1996.9050767.x
Lamb JC et al (2007) Single-gene detection and karyotyping using small-target fluorescence in situ hybridization on maize somatic chromosomes. Genetics 175:1047–1058. https://doi.org/10.1534/genetics.106.065573
Gerhard DS, Kawasaki ES, Bancroft FC, Szabo P (1981) Localization of a unique gene by direct hybridization in situ. Proc Natl Acad Sci USA 78:3755–3759
Harper ME, Ullrich A, Saunders GR (1981) Localization of the human insulin gene to the distal end of the short arm of chromosome 11. Proc Natl Acad Sci USA 78:4458–4460. https://doi.org/10.1073/pnas.78.7.4458
Harper ME, Marselle LM (1985) In situ hybridization: applicabon to gene localization and RNA detection. Can Genet Cytogenet 19:73–80. https://doi.org/10.1016/0165-4608(86)90374-2
Lawrence JB, Villnave CA, Singer RH (1988) Sensitive, high resolution chromatin and chromosome mapping in situ: presence and orientation of two closely integrated copies of EBV in a lymphoma line. Cell 52:51–61. https://doi.org/10.1016/0092-8674(88)90530-2
Henry HQ, Heng JS, Tsui LC (1992) High-resolution mapping of mammalian genes by in situ hybridization to free chromatin. Proc Natl Acad Sci USA 89:9509–9513. https://doi.org/10.1073/pnas.89.20.9509
Lehfer H, Busch W, Martin R, Herrmann RG (1993) Localization of the B-hordein locus on barley chromosomes using fluorescence in situ hybridization. Chromosoma 102:428–432. https://doi.org/10.1007/BF00360408
Jiang J, Gill BS, Wang GL, Ronald PC, Ward DC (1995) Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc Natl Acad Sci USA 92:4487–4491. https://doi.org/10.1073/pnas.92.10.4487
Greilhuber J (1977) Why plant chromosomes do not show G-bands. Theor Appl Genet 50:121–124. https://doi.org/10.1007/BF00276805
Schubert I, Rieger R, Döbel P (1984) G and/or C-bands in plant chromosomes? J Cell Sci 71:11–120
Mehrotra S, Goyal V (2014) Repetitive sequences in plant nuclear DNA: types, distribution, evolution and function. Genom Proteom Bioinform 12:164–171. https://doi.org/10.1016/j.gpb.2014.07.003
Ohmido N, Akiyama Y, Fukui K (1998) Physical mapping of unique nucleotide sequences on identified rice chromosomes. Plant Mol Biol 38:1043–1052. https://doi.org/10.1023/A:1006062905742
Bernini C, Marin- Morales MA (2001) Karyotype analysis in Brachiaria (Poaceae) species. Cytobios 104:157–171
Akiyama Y, Yamada-Akiyama H, Ebina M (2010) Morphological diversity of chromosomes bearing ribosomal DNA loci in Brachiaria species. Grassl Sci 56:217–223. https://doi.org/10.1111/j.1744-697X.2010.00197.x
Nielen S, Almeida LM, Carneiro VT, Araujo AC (2010) Physical mapping of rDNA genes corroborates allopolyploid origin in apomictic Brachiaria brizantha. Sex Plant Reprod 23:45–51. https://doi.org/10.1007/s00497-009-0124-1
Nani TF, Pereira DL, Sobrinho FS, Techio VH (2016) Physical map of repetitive DNA sites in Brachiaria spp.: intravarietal and interspecific polymorphisms. Crop Sci 56:1769–1783. https://doi.org/10.2135/cropsci2015.12.0760 2016.
Santos FC, Guyot R, do Valle CB, Chiari L, Techio VH, Heslop-Harrison P, Vanzela AL (2015) Chromosome distribution and evolution of abundant retrotransposons in plants: gypsy elements in diploid and polyploid Brachiaria forage grasses. Chromosom Res 23:571–582. https://doi.org/10.1007/s10577-015-9492-6
Bennetzen JL et al (2012) Reference genome sequence of the model plant Setaria. Nat Biotechnol 30:555–561. https://doi.org/10.1038/nbt.2196
Peterson DG, Lapitan NLV, Stack SM (1999) Localization of single- and low-copy sequences on tomato synaptonemal complex spreads using fluorescence in situ hybridization (FISH). Genetics 152:427–439
Giussani LM, Cota-Sánchez JH, Zuloaga FO, Kellogg EA (2001) A molecular phylogeny of the grass subfamily Panicoideae (Poaceae) shows multiple origins of C4 photosynthesis. Am J Bot 88:1993–2012. https://doi.org/10.2307/3558427
Schnable JC, Freeling M, Lyons E (2012) Genome-wide analysis of syntenic gene deletion in the grasses. Genome Biol Evol 4:265–277. https://doi.org/10.1093/gbe/evs009
Lyons E, Pedersen B, Kane J, Freeling M (2008) The value of nonmodel genomes and an example using SynMap within CoGe to dissect the hexaploidy that predates the rosids. Trop Plant Biol 1:181–190. https://doi.org/10.1007/s12042-008-9017-y
Wang L, Si Y, Dedow LK, Shao Y, Liu P, Brutnell TP (2011) A low-cost library construction protocol and data analysis pipeline for illumina-based strand-specific multiplex RNA-Seq. PLoS ONE 6/11:e26426. https://doi.org/10.1371/journal.pone.0026426
Grabherr MG et al (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. https://doi.org/10.1038/nbt.1883
Kibbe WA (2007) OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res 35:W43–W46. https://doi.org/10.1093/nar/gkm234
Kato A, Lamb JC, Birchler JA (2004) Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc Natl Acad Sci USA 101:13554–13559. https://doi.org/10.1073/pnas.0403659101
Washburn JD, Schnable JC, Davidse G, Pires JC (2015) Phylogeny and photosynthesis of the grass tribe Paniceae. Am J Bot 102:1–13. https://doi.org/10.3732/ajb.1500222
Lamb JC, Birchler JA (2006) Retroelement genome painting: cytological visualization of retroelement expansions in the genera Zea and Tripsacum. Genetics 173:1007–1021. https://doi.org/10.1534/genetics.105.053165
Silva PIT, Martins AM, Gouvea EG, Pessoa-Filho M, Ferreira ME (2013) Development and validation of microsatellite markers for Brachiaria ruziziensis obtained by partial genome assembly of Illumina single-end reads. Genomics 14:2–9. https://doi.org/10.1186/1471-2164-14-17
Sumner AT (2008) Chromosomes, the karyotype and evolution, in chromosomes: organization and function. Blackwell Science Ltd, Oxford
Paula CMP de, Souza Sobrinho F, Techio VH (2015) Genome constitution and relantioship in Urochloa (Poaceae) species and hybrids. Crop Sci 57:2605–2616. https://doi.org/10.2135/cropsci2017.05.0307
Mendes DV, Boldrini KR, Mendes-Bonato AB, Pagliarini MS, Valle CB do (2006) Cytological evidence of natural hybridization in Brachiaria brizantha Stapf (Gramineae). Bot J Linn Soc 150:441–446. https://doi.org/10.1111/j.1095-8339.2006.00493.x
Leitch IJ, Heslop-Harrison JS (1993) Physical mapping of four sites of 5 s rDNA sequences and one site of the a-amylase-2 gene in barley (Hordeum vulgare). Genome 36:517–523. https://doi.org/10.1139/g93-071
Webb CA, Richter TE, Collins NC, Nicolas M, Trick HN, Pryor T, Hulbert SH (2002) Genetic and molecular characterization of the maize rp3 rust resistance locus. Genetics 162:381–394
Smith SM, Pryor AJ, Hulbert SH (2004) Allelic and haplotypic diversity at the rp1 rust resistance locus of maize. Genetics 167:1939–1947. https://doi.org/10.1534/genetics.104.029371
Woo YM, Hu DW, Larkins BA, Jung R (2001) Genomics analysis of genes expressed in maize endosperm identifies novel seed proteins and clarifies patterns of zein gene expression. Plant Cell 13:2297–2317. https://doi.org/10.1105/tpc.010240
Wu Y, Meeley RB, Cosgrove DJ (2001) Analysis and expression of the alpha-expansin and beta-expansin gene families in maize. Plant Physiol 126:222–232. https://doi.org/10.1104/pp.126.1.222
Abrouk M, Murat F, Pont C, Messing J, Jackson S, Faraut T, Tannier E, Plomion C, Cooke R, Feuillet C, Salse J (2010) Palaeogenomics of plants: synteny-based modelling of extinct ancestors. Trends Plant Sci 15:479–487. https://doi.org/10.1016/j.tplants.2010.06.001
Yu W, Lamb JC, Han F, Birchler JA (2007) Cytological visualization of transposable elements and their transposition pattern in somatic cells of maize. Genetics 175:31–39. https://doi.org/10.1534/genetics.106.064238
Song R, Messing J (2003) Gene expression of a gene family in maize based on noncollinear haplotypes. Proc Natl Acad Sci USA 100:9055–9060. https://doi.org/10.1073/pnas.1032999100
Brunner S, Fengler K, Morgante M, Tingey S, Rafalskia A (2005) Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell 17:343–360. https://doi.org/10.1105/tpc.104.025627
Lai J, Li Y, Messing J, Dooner HK (2005) Gene movement by Helitron transposons contributes to the haplotype variability of maize. Proc Natl Acad Sci USA 102:9068–9073. https://doi.org/10.1073/pnas.0502923102
Richard F, Vogt N, Muleris M, Malfoy B, Dutrillaux B (1994) Increased FISH efficiency using APC probes generated by direct incorporation of labelled nucleotides by PCR. Cytogenet Cell Genet 65:169–171. https://doi.org/10.1159/000133624
Acknowledgements
The authors are thankful to Dr. Ryan Douglas at University of Missouri for helping in designing the centromeric retrotransposons of maize 1 probe (CRM1), and to the Brazilian agencies of research support as Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG), and the program “Science without Borders” of the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nani, T.F., Schnable, J.C., Washburn, J.D. et al. Location of low copy genes in chromosomes of Brachiaria spp.. Mol Biol Rep 45, 109–118 (2018). https://doi.org/10.1007/s11033-018-4144-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-018-4144-5