Abstract
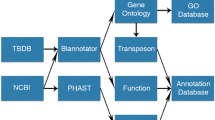
The number of effective drugs for the prevention and control of tuberculosis is very limited. Therefore, high-throughput screening for Mycobacterium tuberculosis drug targets is critical. In addition, determining the essential gene cluster is important for both understanding a survival mechanism and finding novel molecular targets for anti-tuberculosis drugs. In this study, we applied the pathway enrichment method to perform high throughput screening of genes encoding key molecules for potential drug targets for M. tuberculosis. Our results indicated 122 genes that existed in more than three pathways, while four existed in 11 pathways. We predicted 55 genes that are potentially essential genes. Four of them, namely, Rv0363c, Rv0408, Rv0409 and Rv0794c, had the highest probability to be essential genes, and thus further experimental validation is warranted.

Similar content being viewed by others
References
Dutta NK et al (2010) Genetic requirements for the survival of tubercle bacilli in primates. J Infect Dis 201:1743–1752
Cavusoglu C et al (2004) Genotyping of rifampin-resistant Mycobacterium tuberculosis isolates from western Turkey. Ann Saudi Med 24:102–105
Sarkar S, Suresh MR (2011) An overview of tuberculosis chemotherapy—a literature review. J Pharm Pharm Sci 14:148–161
Meena LS, Rajni (2010) Survival mechanisms of pathogenic Mycobacterium tuberculosis H37Rv. The FEBS journal 277:2416–2427
Gerdes S et al (2006) Essential genes on metabolic maps. Curr Opin Biotechnol 17:448–456
Koonin EV (2003) Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat Rev Microbiol 1:127–136
Glass JI et al (2006) Essential genes of a minimal bacterium. Proc Natl Acad Sci USA 103:425–430
Kobayashi K et al (2003) Essential Bacillus subtilis genes. Proc Natl Acad Sci USA 100:4678–4683
Hu W et al (2007) Essential gene identification and drug target prioritization in Aspergillus fumigatus. PLoS Pathog 3:e24
Haselbeck R et al (2002) Comprehensive essential gene identification as a platform for novel anti-infective drug discovery. Curr Pharm Des 8:1155–1172
Giaever G et al (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391
Ji Y et al (2001) Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science 293:2266–2269
Forsyth RA et al (2002) A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol 43:1387–1400
Awasthy D et al (2012) Alanine racemase mutants of Mycobacterium tuberculosis require d-alanine for growth and are defective for survival in macrophages and mice. Microbiology 158:319–327
Ta P et al (2011) Organic hydroperoxide resistance protein and ergothioneine compensate for loss of mycothiol in Mycobacterium smegmatis mutants. J Bacteriol 193:1981–1990
Zeng J et al (2012) A genome-wide regulator-DNA interaction network in the human pathogen Mycobacterium tuberculosis H37Rv. J Proteome Res 11:4682–4692
Tong X et al (2004) Genome-scale identification of conditionally essential genes in E. coli by DNA microarrays. Biochem Biophys Res Commun 322:347–354
Rao M et al (2001) Intracellular pH regulation by Mycobacterium smegmatis and Mycobacterium bovis BCG. Microbiology 147:1017–1024
Deckers-Hebestreit G, Altendorf K (1996) The F0F1-type ATP synthases of bacteria: structure and function of the F0 complex. Annu Rev Microbiol 50:791–824
Rivers EC, Mancera RL (2008) New anti-tuberculosis drugs in clinical trials with novel mechanisms of action. Drug Discovery Today 13:1090–1098
Rivers EC, Mancera RL (2008) New anti-tuberculosis drugs with novel mechanisms of action. Curr Med Chem 15:1956–1967
Godreuil S et al (2007) Genetic diversity and population structure of Mycobacterium tuberculosis in HIV-1-infected compared with uninfected individuals in Burkina Faso. AIDS 21:248–250
Mdluli K, Spigelman M (2006) Novel targets for tuberculosis drug discovery. Curr Opin Pharmacol 6:459–467
Chatterjee D (1997) The mycobacterial cell wall: structure, biosynthesis and sites of drug action. Curr Opin Chem Biol 1:579–588
Zhang XC et al (2011) Cancer nursing research output and topics in the first decade of the 21st century: results of a bibliometric and co-word cluster analysis. Asian Pac J Cancer Prev 12:2055–2058
Acknowledgments
This work was supported by National Natural Science Foundation of China (81101295 and 81071424), Specialized Research Fund for the Doctoral Program of Higher Education of China (20110061120093), China Postdoctoral Science Foundation(20110491311 and 2012T50304),Foundation of Xinjiang Provincial Science & Technology Department(201091148)and Foundation of Jilin Provincial Health Department (2010Z034 and 2011Z049),Bethune special foundation of Jilin University. It was also supported in part by National Basic Research Program of China (973 program, 2011CB512003).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Guangyu Xu and Zhaohui Ni are contributed equally to the work reported here. Guoqing Wang and Fan Li are co-corresponding authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, G., Ni, Z., Shi, Y. et al. Screening essential genes of Mycobacterium tuberculosis with the pathway enrichment method. Mol Biol Rep 41, 7639–7644 (2014). https://doi.org/10.1007/s11033-014-3654-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3654-z