Abstract
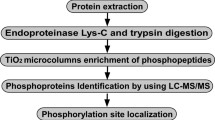
Induction and break of bud dormancy are important features for perennial plants surviving extreme seasonal variations in climate. However, the molecular mechanism of the dormancy regulation, still remain poorly understood. To better understand the molecular basis of poplar bud dormancy, we used a label-free quantitative proteomics method based on nanoscale ultra performance liquid chromatography-ESI-MSE for investigation of differential protein expression during dormancy induction, dormancy, and dormancy break in apical buds of poplar (Populus simonii × P. nigra). Among these identified over 300 proteins during poplar bud dormancy, there are 74 significantly altered proteins, most of which involved in carbohydrate metabolism (22 %), redox regulation (19 %), amino acid transport and metabolism (10 %), and stress response (8 %). Thirty-one of these proteins were up-regulated, five were down-regulated during three phase, and thirty-eight were expressed specifically under different conditions. Pathway analysis suggests that there are still the presence of various physiological activities and a particular influence on photosynthesis and energy metabolism during poplar bud dormancy. Differential expression patterns were identified for key enzymes involved in major metabolic pathways such as glycolysis and the pentose phosphate pathway, thus manifesting the interplay of intricate molecular events in energy generation for new protein synthesis in the dormant buds. Furthermore, there are significant changes present in redox regulation and defense response proteins, for instance in peroxidase and ascorbate peroxidase. Overall, this study provides a better understanding of the possible regulation mechanisms during poplar bud dormancy.





Similar content being viewed by others
Abbreviations
- EMRT:
-
Exact mass and retention time
- MSE :
-
Low/high collision energy MS
- LC:
-
Liquid chromatography
- nano-UPLC:
-
Nanoscale ultra performance LC
- PLGS:
-
ProteinLynx Global-Server
- UPLC:
-
Ultra performance LC
- TCA:
-
Trichloroacetic acid
- BSA:
-
Bovine serum albumin
- DTT:
-
Dithiothreitol
- OEE1:
-
Oxygen-evolving enhancer protein 1
- OEE2:
-
Oxygen-evolving enhancer protein 2
- AdoMetS:
-
S-Adenosylmethionine synthetase
- AdoMet:
-
S-Adenosylmethionine
- LRR:
-
Leucine-rich repeat
- APX:
-
Ascorbate peroxidase
- MDAR:
-
Monodehydroascorbate reductase
- SOD:
-
Superoxide dismutase
- AKR:
-
Aldo/keto reductase
- PDI:
-
Protein disulfide isomerase
References
Arora R, Rowland LJ, Tanino K (2003) Induction and release of bud dormancy in woody perennials: a science comes of age. HortScience 38(5):911–921
Lang GA (1987) Dormancy—a New Universal Terminology. HortScience 22(5):817–820
Horvath DP, Anderson JV, Chao WS, Foley ME (2003) Knowing when to grow: signals regulating bud dormancy. Trends Plant Sci 8(11):534–540
Rohde A, Ruttink T, Hostyn V, Sterck L, Van Driessche K, Boerjan W (2007) Gene expression during the induction, maintenance, and release of dormancy in apical buds of poplar. J Exp Bot 58(15–16):4047–4060
Nitsch J (1957) Growth responses of woody plants to photoperiodic stimuli. Proc Am Soc Hort Sci 70:512–525
Sylven N (1940) Long- and short-day types of Swedish forest trees. Svensk Papperstidning 43:317–324
Allona I, Ramos A, Ibáñez C, Contreras A, Casado R, Aragoncillo C (2008) Review. Molecular control of winter dormancy establishment in trees. Span J Agric Res 6:201–210
Wareing P (1956) Photoperiodism in woody plants. Annu Rev Plant Physiol 7(1):191–214
Howe GT, Hackett WP, Furnier GR, Klevorn RE (1995) Photoperiodic responses of a northern and southern ecotype of black cottonwood. Physiol Planta 93(4):695–708
Li CY, Puhakainen T, Welling A, Vihera-Aarnio A, Ernstsen A, Junttila O, Heino P, Pavla ET (2002) Cold acclimation in silver birch (Betula pendula). Development of freezing tolerance in different tissues and climatic ecotypes. Physiol Planta 116(4):478–488
Nooden L, Weber J (1978) Environmental and hormonal control of dormancy in terminal buds of plants. Dormancy and developmental arrest. Academic Press, New York, pp 222–268
Suttle JC (2000) The role of endogenous hormones in potato tuber dormancy. Dormancy in plants. CAB Publishing, New York, pp 211–226
Liu CC, Liu CF, Wang HX, Shen ZY, Yang CP, Wei ZG (2011) Identification and analysis of phosphorylation status of proteins in dormant terminal buds of poplar. BMC Plant Biol 11
Rohde A, Bhalerao RP (2007) Plant dormancy in the perennial context. Trends Plant Sci 12(5):217–223
Mazzitelli L, Hancock RD, Haupt S, Walker PG, Pont SDA, McNicol J, Cardle L, Morris J, Viola R, Brennan R, Hedley PE, Taylor MA (2007) Co-ordinated gene expression during phases of dormancy release in raspberry (Rubus idaeus L.) buds. J Exp Bot 58(5):1035–1045
Mathiason K, He D, Grimplet J, Venkateswari J, Galbraith D, Or E, Fennell A (2009) Transcript profiling in Vitis riparia during chilling requirement fulfillment reveals coordination of gene expression patterns with optimized bud break. Funct Integr Genomic 9(1):81–96
Jimenez S, Li ZG, Reighard GL, Bielenberg DG (2010) Identification of genes associated with growth cessation and bud dormancy entrance using a dormancy-incapable tree mutant. Bmc Plant Biol 10
Liu CC, Lu TC, Li HH, Wang HX, Liu GF, Ma L, Yang CP, Wang BC (2010) Phosphoproteomic identification and phylogenetic analysis of ribosomal P-proteins in Populus dormant terminal buds. Planta 231(3):571–581
Muthalif MM, Rowland LJ (1994) Identification of dehydrin-like proteins responsive to chilling in floral buds of blueberry (Vaccinium, Section Cyanococcus). Plant Physiol 104(4):1439–1447
Renaut J, Lutts S, Hoffmann L, Hausman JF (2004) Responses of poplar to chilling temperatures: proteomic and physiological aspects. Plant Biol 6(1):81–90
Bassett CL, Wisniewski ME, Artlip TS, Richart G, Norelli JL, Farrell RE (2009) Comparative expression and transcript initiation of three peach dehydrin genes. Planta 230(1):107–118
Palonen P, Buszard D, Donnelly D (2000) Changes in carbohydrates and freezing tolerance during cold acclimation of red raspberry cultivars grown in vitro and in vivo. Physiol Planta 110(3):393–401
Rinne PLH, van der Schoot C (1998) Symplasmic fields in the tunica of the shoot apical meristem coordinate morphogenetic events. Development 125(8):1477–1485
Rinne PLH, Kaikuranta PM, van der Schoot C (2001) The shoot apical meristem restores its symplasmic organization during chilling-induced release from dormancy. Plant J 26(3):249–264
Nir G, Shulman Y, Fanberstein L, Lavee S (1986) Changes in the activity of catalase (EC 1.11. 1.6) in relation to the dormancy of grapevine (Vitis vinifera L.) buds. Plant Physiol 81(4):1140
Or E, Vilozny I, Fennell A, Eyal Y, Ogrodovitch A (2002) Dormancy in grape buds: isolation and characterization of catalase cDNA and analysis of its expression following chemical induction of bud dormancy release. Plant Sci 162(1):121–130
Wang SY, Jiao HJ, Faust M (1991) Changes in ascorbate, glutathione, and related enzyme-activities during thidiazuron-induced bud break of apple. Physiol Planta 82(2):231–236
Halaly T, Pang XQ, Batikoff T, Crane O, Keren A, Venkateswari J, Ogrodovitch A, Sadka A, Lavee S, Or E (2008) Similar mechanisms might be triggered by alternative external stimuli that induce dormancy release in grape buds. Planta 228(1):79–88
Shen Z, Li P, Ni RJ, Ritchie M, Yang CP, Liu GF, Ma W, Liu GJ, Ma L, Li SJ, Wei ZG, Wang HX, Wang BC (2009) Label-free quantitative proteomics analysis of etiolated maize seedling leaves during greening. Mol Cell Proteomics 8(11):2443–2460
Silva JC, Denny R, Dorschel C, Gorenstein MV, Li GZ, Richardson K, Wall D, Geromanos SJ (2006) Simultaneous qualitative and quantitative analysis of the Escherichia coli proteome—a sweet tale. Mol Cell Proteomics 5(4):589–607
Silva JC, Gorenstein MV, Li GZ, Vissers JPC, Geromanos SJ (2006) Absolute quantification of proteins by LCMSE—a virtue of parallel MS acquisition. Mol Cell Proteomics 5(1):144–156
Zhou YL, Dong SL, Nie SQ (1986) Heilongjiang sylva. Heilongjiang science and technology press, Harbin, p118
Yang CP, Liu GF, Liang HW, Zhang H (2001) Study on the transformation of Populus simonii × P. nigra with salt resistance gene Bet-A. Sci Silvae Sinicae 37:34–38
Jonsson TH (2006) Terminal bud failure of black cottonwood (Populus trichocarpa) exposed to salt-laden winter storms. Tree Physiol 26(7):905–914
Way DA (2011) Tree phenology responses to warming: spring forward, fall back? Tree Physiol 31(5):469–471
Washburn MP, Wolters D, Yates JR (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol 19(3):242–247
Meng YY, Liu F, Pang CY, Fan SL, Song MZ, Wang D, Li WH, Yu SX (2011) Label-free quantitative proteomics analysis of cotton leaf response to nitric oxide. J Proteome Res 10(12):5416–5432
Silva JC, Denny R, Dorschel CA, Gorenstein M, Kass IJ, Li GZ, McKenna T, Nold MJ, Richardson K, Young P, Geromanos S (2005) Quantitative proteomic analysis by accurate mass retention time pairs. Anal Chem 77(7):2187–2200
Hughes MA, Silva JC, Geromanos SJ, Townsend CA (2006) Quantitative proteomic analysis of drug-induced changes in mycobacteria. J Proteome Res 5(1):54–63
Li GZ, Vissers JPC, Silva JC, Golick D, Gorenstein MV, Geromanos SJ (2009) Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics 9(6):1696–1719
Vissers JPC, Langridge JI, Aerts JMFG (2007) Analysis and quantification of diagnostic serum markers and protein signatures for Gaucher disease. Mol Cell Proteomics 6(5):755–766
Kopp RF, Abrahamson LP, White EH, Volk TA, Nowak CA, Fillhart RC (2001) Willow biomass production during ten successive annual harvests. Biomass Bioenerg 20(1):1–7
Oja V, Bichele I, Huve K, Rasulov B, Laisk A (2004) Reductive titration of photosystem I and differential extinction coefficient of P700(+) at 810–950 nm in leaves. Biochim Biophys Acta 1658(3):225–234
Whitelegge JP, Koo D, Diner BA, Domian I, Erickson JM (1995) Assembly of the photosystem II oxygen-evolving complex is inhibited in psbA site-directed mutants of Chlamydomonas reinhardtii—Aspartate-170 of the D1 polypeptide. J Biol Chem 270(1):225–235
Mayfield SP, Bennoun P, Rochaix JD (1987) Expression of the nuclear encoded Oee1 protein is required for oxygen evolution and stability of photosystem II particles in Chlamydomonas reinhardtii. EMBO J 6(2):313–318
Miyao M, Murata N (1989) The mode of binding of 3 extrinsic proteins of 33 kDa, 23 kDa and 18 kDa in the photosystem II complex of spinach. Biochim Biophys Acta 977(3):315–321
Ghanotakis DF, Yocum CF (1990) Photosystem II and the oxygen-evolving complex. Annu Rev Plant Phys 41:255–276
Szegő D, Kósa E, Horváth E (2007) Role of S-methylmethionine in the plant metabolism. Acta Agron Hung 55(4):491–508
Ho P, Kong K, Chan Y, Tsang J, Wong J (2007) An unusual S-adenosylmethionine synthetase gene from dinoflagellate is methylated. BMC Mol Biol 8(1):87
Newman EB, Budman LI, Chan EC, Greene RC, Lin RT, Woldringh CL, D’Ari R (1998) Lack of S-adenosylmethionine results in a cell division defect in Escherichia coli. J Bacteriol 180(14):3614–3619
Ben-Haj-Salah H, Tardieu F (1995) Temperature affects expansion rate of maize leaves without change in spatial distribution of cell length (analysis of the coordination between cell division and cell expansion). Plant Physiol 109(3):861–870
Bergervoet JHW, Jing HC, Van Den Hout JWE, Delmondez de Castro R, Kunneman BPAM, Bino RJ, Groot SPC (1999) Expression of beta-tubulin during dormancy induction and release in apical and axillary buds of five woody species. Physiol Planta 106(2):238–245
Nordin K, Vahala T, Palva ET (1993) Differential expression of two related, low-temperature-induced genes in Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 21(4):641–653
Lin CT, Thomashow MF (1992) A cold-regulated Arabidopsis gene encodes a polypeptide having potent cryoprotective activity. Biochem Bioph Res Co 183(3):1103–1108
Hon WC, Griffith M, Chong PL, Yang DSC (1994) Extraction and isolation of antifreeze proteins from winter rye (Secale-Cereale L.) leaves. Plant Physiol 104(3):971–980
Grudkowska M, Zagdanska B (2004) Multifunctional role of plant cysteine proteinases. Acta Biochim Pol 51(3):609–624
Meyer K, Keil M, Naldrett MJ (1999) A leucine-rich repeat protein of carrot that exhibits antifreeze activity. FEBS Lett 447(2–3):171–178
Johnson R, Narvaez J, An G, Ryan C (1989) Expression of proteinase inhibitors I and II in transgenic tobacco plants: effects on natural defense against Manduca sexta larvae. Proc Natl Acad Sci USA 86(24):9871
Thomas S, Mooney PFJ, Burrell MM, Fell DA (1997) Metabolic control analysis of glycolysis in tuber tissue of potato (Solanum tuberosum): explanation for the low control coefficient of phosphofructokinase over respiratory flux. Biochem J 322:119–127
Wurtele ES, Nikolau BJ (1986) Enzymes of glucose oxidation in leaf tissues: the distribution of the enzymes of glycolysis and the oxidative pentose phosphate pathway between epidermal and mesophyll tissues of C3-plants and epidermal, mesophyll, and bundle sheath tissues of C4-plants. Plant Physiol 82(2):503
Kato-Noguchi H (2007) Low temperature acclimation to chilling tolerance in rice roots. Plant Growth Regul 51(2):171–175
De Vries FWTP (1975) The cost of maintenance processes in plant cells. Ann Bot 39(1):77–92
Hachiya T, Terashima I, Noguchi K (2007) Increase in respiratory cost at high growth temperature is attributed to high protein turnover cost in Petunia × hybrida petals. Plant Cell Environ 30(10):1269–1283
Noguchi K, Nakajima N, Terashima I (2001) Acclimation of leaf respiratory properties in Alocasia odora following reciprocal transfers of plants between high- and low-light environments. Plant Cell Environ 24(8):831–839
Nelson EA, Dickson RE (1981) Accumulation of food reserves in cottonwood stems during dormancy induction. Can J Forest Res 11(1):145–154
Colmer TD, Epstein E, Dvorak J (1995) Differential solute regulation in leaf blades of various ages in salt-sensitive wheat and a salt-tolerant wheat x Lophopyrum elongatum (Host) A. Love amphiploid. Plant Physiol 108(4):1715–1724
Chinnusamy V, Zhu JK, Sunkar R (2010) Gene regulation during cold stress acclimation in plants. Plant Stress Tolerance: Methods and Protocols 639:39–55
Korkmaz A, Dufault RJ (2001) Developmental consequences of cold temperature stress at transplanting on seedling and field growth and yield. II. Muskmelon. J Am Soc Hortic Sci 126(4):410–413
Dat J, Vandenabeele S, Vranova E, Van Montagu M, Inzé* D, Van Breusegem F (2000) Dual action of the active oxygen species during plant stress responses. Cell Mol Life Sci 57(5):779–795
Bailly C (2004) Active oxygen species and antioxidants in seed biology. Seed Sci Res 14(2):93–108
Pastori GM, Foyer CH (2002) Common components, networks, and pathways of cross-tolerance to stress. The central role of “redox” and abscisic acid-mediated controls. Plant Physiol 129(2):460–468
Foyer CH, Noctor G (2003) Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol Planta 119(3):355–364
Kocsy G, Galiba G, Brunold C (2001) Role of glutathione in adaptation and signalling during chilling and cold acclimation in plants. Physiol Planta 113(2):158–164
Wang SY, Jiao HJ, Faust M (1991) Changes in ascorbate, glutathione, and related enzyme activities during thidiazuron-induced bud break of apple. Physiol Planta 82(2):231–236
Wang SY, Faust M (1994) Changes in the antioxidant system associated with budbreak in anna’apple (Malus domestica Borkh.) buds. J Am Soc Hortic Sci 119(4):735–741
Perez FJ, Lira W (2005) Possible role of catalase in post-dormancy bud break in grapevines. J Plant Physiol 162(3):301–308
Bi YD, Wei ZG, Shen Z, Lu TC, Cheng YX, Wang BC, Yang CP (2011) Comparative temporal analyses of the Pinus sylvestris L. var. mongolica litv. apical bud proteome from dormancy to growth. Mol Biol Rep 38(2):721–729
Bartels D (2001) Targeting detoxification pathways: an efficient approach to obtain plants with multiple stress tolerance? Trends Plant Sci 6(7):284–286
Schultz-Norton JR, McDonald WH, Yates JR, Nardulli AM (2006) Protein disulfide isomerase serves as a molecular chaperone to maintain estrogen receptor α structure and function. Mol Endocrinol 20(9):1982–1995
Wells WW, Xu DP, Yang YF, Rocque PA (1990) Mammalian thioltransferase (Glutaredoxin) and protein disulfide isomerase have dehydroascorbate reductase-activity. J Biol Chem 265(26):15361–15364
Liu JJ, Ekramoddoullah AKM (2006) The family 10 of plant pathogenesis-related proteins: their structure, regulation, and function in response to biotic and abiotic stresses. Physiol Mol Plant Pathol 68(1):3–13
Hon WC, Griffith M, Mlynarz A, Kwok YC, Yang DSC (1995) Antifreeze proteins in winter rye are similar to pathogenesis-related proteins. Plant Physiol 109(3):879–889
Gaudet DA, Laroche A, Frick M, Davoren J, Puchalski B, Ergon A (2000) Expression of plant defence-related (PR-protein) transcripts during hardening and dehardening of winter wheat. Physiol Mol Plant P 57(1):15–24
Acknowledgments
The authors acknowledge grant support from the National Basic Research Priorities Program (Grant No. 2009CB119102).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ning, DL., Liu, CC., Liu, JW. et al. Label-free quantitative proteomics analysis of dormant terminal buds of poplar. Mol Biol Rep 40, 4529–4542 (2013). https://doi.org/10.1007/s11033-013-2548-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-013-2548-9