Abstract
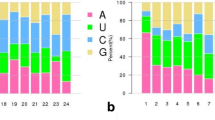
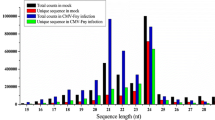
Presence of selected tomato (Solanum lycopersicon) microRNAs (sly-miRNAs) was validated and their expression profiles established in roots, stems, leaves, flowers and fruits of tomato variety Jiangshu14 by quantitative RT-PCR (qRT-PCR). In addition conservation characteristics these sly-miRNAs were analyzed and target genes predicted bioinformatically. Results indicate that some of these miRNAs are specific to tomato while most are conserved in other plant species. Predicted sly-miRNA targets genes were shown to be targeted by either by a single or more miRNAs and are involved in diverse processes in tomato plant growth and development. All the 36 miRNAs were present in the cDNA of mixed tissues and qRT-PCR revealed that some of these sly-miRNAs are ubiquitous in tomato while others have tissue-specific expression. The experimental validation and expression profiling as well target gene prediction of these miRNAs in tomato as done in this study can add to the knowledge on the important roles played by these sly-miRNAs in the growth and development, environmental stress tolerance as well as pest and disease resistance in tomatoes and related species. In addition these findings broaden the knowledge of small RNA-mediated regulation in S. lycopersicon. It is recommended that experimental validation of the target genes be done so as to give a much more comprehensive information package on these miRNAs in tomato and specifically in the selected variety.







Similar content being viewed by others
References
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854
Wightman B, Ha I, Ruvkun G (1993) Posttranscriptional regulation of the heterochronic gene Lin-14 by Lin-4 mediates temporal pattern-formation in C. elegans. Cell 75:855–862
Krol J, Loedige I, Filipowicz W (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11:597–610
Chekulaeva M, Filipowicz W (2009) Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol 21:452–460
Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JDG (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312:436–439
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Mallory AC, Vaucheret H (2004) MicroRNAs: something important between the genes. Curr Opin Plant Biol 7:120–125
Carrington JC, Ambros V (2003) Role of microRNAs in plant and animal development. Science 301:336–338
Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57:19–53
Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MC (2004) MicroRNA-mediated repression of rolled leaf specifies maize leaf polarity. Nature 428:84–88
Xu L, Yang L, Hang H (2007) Transcriptional, post-transcriptional and post-translational regulations of gene expression during leaf polarity formation. Cell Res 17:512–519
Nag A, King S, Jack T (2009) miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. PNAS 106:22534–22539
Nag A, Jack T (2010) Chapter twelve-sculpting the flower; the role of microRNAs in flower development. Curr Top Dev Biol 91:349–378
Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16:2001–2019
Wang XJ, Reyes JL, Chua NHT (2004) Gaasterland, prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol 5:R65
Kim HJ, Baek KH, Lee BW, Choi D, Hur CG (2011) In-silico identification and characterization of microRNAs and their putative target genes in Solanaceae plants. Genome 54(2):91–98
Zhang YP, Yu ML, Yu HP, Han J, Song CN, Ma RJ, Fang JG (2011) Computational identification of microRNAs in peach expressed sequence tags and validation of their precise sequences by miR-RACE. Mol Biol Rep. doi:10.1007/s11033-011-0944-6
Zeng CY, Wang WQ, Zheng Y, Chen X, Bo WP, Song S, Zhang WX, Peng M (2010) Conservation and divergence of microRNAs and their functions in Euphorbiaceous plants. Nucleic Acids Res 38:981–995
Yu HP, Song CN, Jia QD, Wang C, Li F, Nicholas KK, Zhang XY, Fang JG (2010) Computational identification of microRNAs in apple expressed sequence tags and validation of their precise sequences by miR-RACE. Physiol Plant 141:56–70
Song CN, Fang JG, Li XY, Liu H, Chao CT (2009) Identification and characterization of 27 conserved microRNAs in citrus. Planta 230:671–685
Carra A, Mica E, Gambino G, Pindo M, Moser C, Enrico MP, Schubert A (2009) Cloning and characterization of small non-coding RNAs from grape. Plant J 59:750–763
Yin ZJ, Li CH, Han XL, Shen FF (2008) Identification of conserved microRNAs and their target genes in tomato (Lycopersicon esculentum). Gene 414:60–66
Zhang BH, Pan XP, Stellwag EJ (2008) Identification of soybean microRNAs and their targets. Planta 229:161–182
Gleave AP, Ampomah-Dwamena C, Berthold S, Dejnoprat S, Karunairetnam S, Nain B, Wang YY, Crowhurst RN, MacDiarmid RM (2008) Identification and characterisation of primary microRNAs from apple (Malus domestica cv. Royal Gala) expressed sequence tags. Tree Genet Genomes 4:343–358
Zhang BH, Wang QL, Wang KB, Pan XP, Liu F, Guo TL, Cobb GP, Anderson TA (2007) Identification of cotton microRNAs and their targets. Gene 397:26–37
Xie FL, Huang SQ, Guo K, Xiang AL, Zhu YY, Nie L, Yang ZM (2007) Computational identification of novel microRNAs and targets in Brassica napus. FEBS Lett 581:1464–1474
Qiu CX, Xie FL, Zhu YY, Guo K, Huang SQ, Nie L, Yang ZM (2007) Computational identification of microRNAs and their targets in Gossypium hirsutum expressed sequence tags. Gene 395:49–61
Zhang BH, Pan XP, Anderson TA (2006) Identification of 188 conserved maize microRNAs and their targets. FEBS Lett 580:3753–3762
Leonardi C, Ambrosino P, Esposito F, Fogliano V (2000) Antioxidant activity and carotenoid and tomatine contents in different typologies of fresh ‘consumption tomatoes. J Agric Food Chem 48:4723–4727
Mohorianu I, Schwach F, Jing R, Lopez-Gomollon S, Moxon S, Szittya G, Sorefan K, Moulton V, Dalmay T (2011) Profiling of short RNAs during fleshy fruit development reveals stage-specific sRNAome expression patterns. Plant J 67(2):232–246
Moxon S, Jing R, Szittya G, Schwach F, Pilcher RL, Moulton V, Dalmay T (2008) Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res 18:1602–1609
Itaya A, Bundschuh R, Archual AJ, Joung JG, Fei Z, Dai X, Zhao PX, Tang Y, Nelson RS, Ding B (2008) Small RNAs in tomato fruit and leaf development. Biochim Biophys Acta 1779(2):99–107
Pilcher RL, Moxon S, Pakseresht N, Moulton V, Manning K, Seymour G, Dalmay T (2007) Identification of novel small RNAs in tomato (Solanum lycopersicum). Planta 226(3):709–717
Zuo JH, Wang YX, Liu HP, Ma YZ, Ju Z, Zhai BQ, Fu DQ, Zhu Y, Luo YB, Zhu BZ (2011) MicroRNAs in tomato plants. Life Sci 54(7):599–605
Li YF, Zheng Y, Addo-Quaye C, Zhang L, Saini A, Jagadeeswaran G, Axtell MJ, Zhang WX, Sunker R (2010) Transcriptome-wide identification of microRNA targets in rice. Plant J 62:742–759
Mallory AC, Bartel DP, Bartel B (2005) MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17:1360–1375
Floyd SK, Bowman JL (2004) Gene regulation: ancient microRNA target sequences in plants. Nature 428:485–486
Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425:257–263
Wang C, Han J, Liu CH, Korir NK, Kayesh E, Shangguan L, Li XY, Fang JG (2012) Identification of microRNAs from Amur grape (Vitis amurensis Rupr.) by deep sequencing and analysis of microRNA variations with bioinformatics. BMC Genomics 13:122
Song CN, Fang JG, Wang C, Guo L, Nicholas KK, Ma Z (2010) miR-RACE, a new efficient approach to determine the precise sequences of computationally identified trifoliate orange (Poncirus trifoliata) microRNAs. PLoS ONE 5:e10861
Song CN, Jia QD, Fang JG, Li F, Wang C, Zhang Z (2010) Computational identification of citrus microRNAs and target analysis in citrus expressed sequence tags. Plant Biol 12:927–934
Sunkar R, Jagadeeswaran G (2008) In silico identification of conserved microRNAs in a large number of diverse plant species. BMC Plant Biol 8:37
Zhang BH, Pan X, Cannon CH, Cobb GP, Anderson TA (2006) Conservation and divergence of plant microRNA genes. Plant J 46:243–259
Sunkar R, Zhou XF, Zheng Y, Zhang WX, Zhu JK (2008) Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol 8:25
Bonnet E, Wuyts J, Rouze′ P, Van de Peer Y (2004) Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. PNAS 101:11511–11516
Griffiths-Jones S, Saini HK, Van Dongen S, Enrigh AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36(suppl 1):D154–D158
Adai A, Johnson C, Mlotshwa S, Archer-Evans S, Manocha V, Vance V, Sundaresan V (2005) Computational prediction of miRNAs in Arabidopsis thaliana. Genome Res 15:78–91
Wang C, Shangguan LF, Nicholas KK, Wang XC, Han J, Song CN, Fang JG (2011) Characterization of microRNAs identified in a table grapevine cultivar with validation of computationally predicted grapevine miRNAs by miR-RACE. PLoS ONE 6:e21259
Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8:517–527
Shi R, Chiang VL (2005) Facile means for quantifying microRNA expression by real-time PCR. Biotechniques 39:519–525
Ramakers C, Ruijter JM, Deprez RH, Moorman AFM (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339:62–66
Fu HJ, Tie Y, Xu CW, Zhang ZY, Zhu J, Shi YX, Jiang H, Sun ZX, Zheng XF (2005) Identification of human fetal liver miRNAs by a novel method. FEBS Lett 579:3849–3854
Altschul SF, Madden TL, Schäffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Tang G (2010) Plant microRNAs: an insight into their gene structures and evolution. Semin Cell Dev Biol 21:782–789
Ambros V (2004) The functions of animal microRNAs. Nature 431:350–355
Palatnik JF, Wollmann H, Schommer C, Schwab R, Boisbouvier J, Rodriguez R, Warthmann N, Allen E, Dezulian T, Husan D, Carrington JC, Weigel D (2007) Sequence and expression differences underlie functional specialization of arabidopsis microRNAs miR159 and miR319. Dev Cell 13:115–125
Mallory AC, Bouché N (2008) MicroRNA-directed regulation: to cleave or not to cleave. Trends Plant Sci 13:359–367
Llave C, Xie ZX, Kasschau KD, Carrington JC (2002) Cleavage of scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297:2053–2056
Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant micro-RNAs and their targets, including a stress-induced miRNA. Mol Cell 14:787–799
Zhang BH, Wang QL, Wang KB, Pan XP, Liu F, Guo TL, Cobb GP, Anderson TA (2006) Computational identification of microRNAs and their targets. Comput Biol Chem 30:395–407
Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110:513–520
Dalmay T (2010) Short RNAs in tomato. J Integr Plant Biol 52(4):388–392
Zhang BH, Pan XP, Cobb GP, Anderson TA (2006) Plant microRNA: a small regulatory molecule with big impact. Dev Biol 289:3–16
Sunkar R, Girke T, Jain PK, Zhu JK (2005) Cloning and characterization of microRNAs from rice. Plant Cell 17:1397–1411
Chen XM (2008) MicroRNA metabolism in plants. Curr Top Microbiol Immunol 320:117–136
Acknowledgments
This work was supported by A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, the NCET Program of China (Grant No. NCET-08-0796), and the Fundamental Research Funds for the Central Universities (Grant No. KYJ200909).
Author information
Authors and Affiliations
Corresponding author
Appendix
Rights and permissions
About this article
Cite this article
Korir, N.K., Li, X., Xin, S. et al. Characterization and expression profiling of selected microRNAs in tomato (Solanum lycopersicon) ‘Jiangshu14’. Mol Biol Rep 40, 3503–3521 (2013). https://doi.org/10.1007/s11033-012-2425-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-2425-y