Abstract
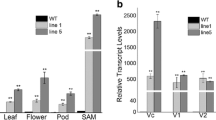
A MADS box gene AGL20/SOC1 is a main integrator in Arabidopsis flowering pathway whose structure and function are highly conserved in many plant species. A soybean MADS box gene GmGAL1 (G lycine max A GAMOUS L ike 1) as a homolog of AGL20/SOC1, was cloned from soybean cultivar Kennong18 and its function was investigated in transgenic Arabidopsis lines. Sequence comparisons showed that the closest homolog gene to GmGAL1 is AGL20/SOC1 in Arabidopsis and VuSOC1 in Vigna unguiculata. We investigated the expression level of GmGAL1 using quantitative real-time PCR, and the result showed that GmGAL1 was regulated by a circadian mechanism and its expression oscillated at a cycle of 24 h. The expression level of GmGAL1 was fluctuated in diverse tissues/organs and developmental stages. Considering its expression can be detected in different tissues throughout the life cycle and its protein localized in cytoplasm in Arabidopsis protoplasm, we proposed that GmGAL1 may be a multifunctional gene in the context of the soybean development. Ectopic expression of GmGAL1 in Arabidopsis enhanced flowering under long-day condition and partially rescued soc1 late flowering type.







Similar content being viewed by others
References
Bodt DS, Raes J, Van de Peer Y, Theissen G (2003) And then there were many: MADS goes genomic. Trends Plant Sci 8:475–483
Foo E, Ross JJ, Davies NW, Reid JB, Weller JL (2006) A role for ethylene in the phytochrome-mediated control of vegetative development. Plant J 46:911–921
Irish V (2003) The evolution of floral homeotic gene function. Bioessays 25:637–646
Kater M, Dreni L, Colombo L (2006) Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J Exp Bot 57:3433–3444
Messenguy F, Dubois E (2003) Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 316:1–21
Rijpkema A, Gerats T, Vandenbussche M (2007) Evolutionary complexity of MADS complexes. Curr Opin Plant Biol 10:32–38
Robles P, Pelaz S (2005) Flower and fruit development in Arabidopsis thaliana. Int J Dev Biol 49:33–43
Saedler H, Huijser P (1993) Molecular biology of flower development in Antirrhinum majus (snapdragon). Gene 135:239–243
Hepworth S, Valverde Ravenscroft FD, Mouradov A, Coupland G (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J 21:4327–4337
Poduska B, Humphrey T, Redweik A, Grbic V (2003) The synergistic activation of FLOWERING LOCUS C by FRIGIDA and a new flowering gene AERIAL ROSETTE 1 underlies a novel morphology in Arabidopsis. Genetics 163:1457–1465
Rouse DT, Sheldon CC, Bagnall DJ, Peacock WJ, Dennis ES (2002) FLC, a repressor of flowering, is regulated by genes in different inductive pathways. Plant J 29:183–191
Swarup K, Alonso-Blanco C, Lynn JR, Michaels SD, Amasino RM, Koornneef M, Millar AJ (1999) Natural allelic variation identifies new genes in the Arabidopsis circadian system. Plant J 20:67–77
Sessions A, Yanofsky MF, Weigel D (2000) Cell–cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science 289:779–782
Wagner D, Sablowski RWM, Meyerowitz EM (1999) Transcriptional activation of APETALA1 by LEAFY. Science 285:582–584
Tapia-López R, García-Ponce B, Dubrovsky JG, Garay-Arroyo A, Pérez-Ruíz RV, Kim SH, Acevedo F, Pelaz S, Alvarez-Buylla ER (2008) An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol 46:1182–1192
Kim S, Kim SR, An CS, Hong YN, Lee KW (2001) Constitutive expression of rice MADS box gene using seed explants in hot pepper (Capsicum annuum L.). Mol Cells 12:221–226
Lee S, Kim J, Han JJ, Han MJ, An G (2004) Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J 38:754–764
Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35:613–623
Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288:1613–1616
Sheldon C, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11:445–458
Borner R, Kampmann G, Chandler J, Gleibner R, Wisman E, Apel K, Melzer S (2000) A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J 24:519–599
Lee Jungeun LI (2010) Regulation and function of SOC1, a flowering pathway integrator. J Exp Bot 61:2247–2254
Liu C, Xi WY, Shen LS, Tan CP, Yu H (2009) Regulation of floral patterning by flowering time genes. Dev Cell 16:711–722
Decroocq V, Zhu XM, Kauffman M, Kyozuka J, Peacock WJ, Dennis ES, Llewellyn DJ (1999) A TM3-like MADS-box gene from Eucalyptus expressed in both vegetative and reproductive tissues. Gene 228:155–160
Tadege M, Sheldon CC, Helliwell CA, Upadhyaya NM, Dennis ES, Peacock WJ (2003) Reciprocal control of flowering time by OsSOC1 in transgenic Arabidopsis and by FLC in transgenic rice. Plant Biotechnol J1:361–369
Heuer S, Hansen S, Bantin J, Brettschneider R, Kranz E, Lorz H, Dresselhaus T (2001) The maize MADS box gene ZmMADS3 affects node number and spikelet development and is co-expressed with ZmMADS1 during flower development, in egg cells, and early embryogenesis. Plant Physiol 127:33–45
Nakamura T, Song IJ, Fukuda T, Yokoyama J, Maki M, Ochiai T, Kameya T, Kanno A (2005) Characterization of TrcMADS1 gene of Trillium camtschatcense (Trilliaceae) reveals functional evolution of the SOC1/TM3-like gene family. J Plant Res 118:229–234
Ferrario S, Busscher J, Franken J, Gerats T, Vandenbussche M, Angenent GC, Immink RG (2004) Ectopic expression of the petunia MADS box gene UNSHAVEN accelerates flowering and confers leaflike characteristics to floral organs in a dominant-negative manner. Plant Cell 16:1490–1505
Zhang Q, Li HY, Li R, Hu RB, Fan CM, Chen FL, Wang ZH, Liu Xu, Fu YF, Lin CT (2008) Association of the circadian rhythmic expression of GmCRY1a with a latitudinal cline in photoperiodic flowering of soybean. Proc Natl Acad Sci USA 105:21028–21033
Xu JH, Zhong XF, Zhang QZ, Li HY (2010) Overexpression of the GmGAL2 gene accelerates flowering in Arabidopsis. Plant Mol Biol Rep 28:704–711
Steven J, Andrew FB (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Melzer S, Lens F, Gennen J, Vanneste S, Rohde A, Beeckman T (2008) Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat Genet 40:1489–1492
Cseke LJZJ, Podila GK (2003) Characterization of PTM5 in aspen trees: a MADS-box gene expressed during woody vascular development. Gene 318:55–67
Tan F, Swain SM (2007) Functional characterization of AP3, SOC1 and WUS homologues from citrus (Citrus sinensis). Physiol Plantarum 131:481–495
Liu H, Wang HG, Gao PF, Xv JH, Xv TD, Wang JS, Wang BL, Lin CT, Fu YF (2009) Analysis of clock gene homologs using unifolioates as target organs in soybean (Glyciine max). J Plant Physiol 166:278–289
Lu SX, Knowles SM, Andronis C, Ong MS, Tobin EM (2009) CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol 150:834–843
Lee J, Oh M, Park H, Lee I (2008) SOC1 translocated to the nucleus by interaction with AGL24 directly regulates LEAFY. Plant J 55:832–843
Acknowledgments
This work was supported in part by the Chinese national “863” Program (2006AA10Z107), Ministry of Agriculture, major projects transgenic (2009ZX08004-010B) and the National Natural Science Foundation of China (Grant No. 31171352).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Zhong, X., Dai, X., Xv, J. et al. Cloning and expression analysis of GmGAL1, SOC1 homolog gene in soybean. Mol Biol Rep 39, 6967–6974 (2012). https://doi.org/10.1007/s11033-012-1524-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-1524-0