Abstract
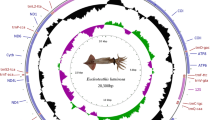
The mitochondrial genome sequences were determined for two species of nemerteans, Cephalothrix sp. (15,800 bp sequenced, near-complete) and Paranemertes cf. peregrina (14,558 bp, complete). As seen in most metazoans, the genomes encode 13 protein, 2 ribosomal RNA and 22 transfer RNA genes. The nucleotide composition is strongly biased toward A and T, as is typical for metazoan mtDNAs. There is also a significant strand skew in the distribution of these nucleotides, with the coding strand being richer in T than A and in G than C. All genes are transcribed in the same direction except for trnP and trnT, which is consistent with that reported for Cephalothrix hongkongiensis and Lineus viridis. Gene arrangement of Cephalothrix sp. is identical to that of C. hongkongiensis, while in P. cf. peregrina it is similar to L. viridis, but differs significantly from the three Cephalothrix species in the position of four protein-coding genes and seven tRNAs. Some protein-coding genes have 3′ end stem-loop structures, which may allow mRNA processing without flanking tRNAs. The major non-coding regions observed in the two genomes with potential to form stem-loop structures may be involved in the initiation of replication or transcription. The average Ka/Ks values, varying from 0.12 to 0.89, are markedly different among the 13 mitochondrial protein-coding genes, suggesting that there may exist different selective pressure among mitochondrial genes of nemerteans.








Similar content being viewed by others
Abbreviations
- atp6 and atp8:
-
ATP synthase subunits 6 and 8
- cob :
-
Cytochrome b
- cox1–3:
-
Subunits I–III of cytochrome c oxidase
- nad1–6 and nad4L:
-
NADH dehydrogenase subunits 1–6 and 4L
- rrnL and rrnS:
-
The large and small subunits of ribosomal RNA
- trnX :
-
Transfer RNA molecules with corresponding amino acids denoted by the one-letter code and codon indicated in parentheses (xxx) when necessary
- DHU:
-
Dihydrouridine loop
- mtDNA:
-
Mitochondrial DNA
- NC:
-
Non-coding region
- PCR:
-
Polymerase chain reaction
- kb:
-
Kilobase
- bp:
-
Base pair
- R:
-
A/G
- Y:
-
C/T
- W:
-
A/T
- K:
-
G/T
- N:
-
A/G/C/T
References
Boore JL (1999) Animal mitochondrial genomes. Nucleic Acids Res 27:1767–1780
Shadel GS, Clayton DA (1997) Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem 66:409–435. doi:10.1146/annurev.biochem.66.1.409
Boore JL, Brown WM (1998) Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Curr Opin Genet Dev 8:668–674. doi:10.1016/S0959-437X(98)80035-X
Boore JL, Macey JR, Medina M (2005) Sequencing and comparing whole mitochondrial genomes of animals. Methods Enzymol 395:311–348. doi:10.1016/S0076-6879(05)95019-2
Kajihara H, Chernyshev AV, Sun SC, Sundberg P, Crandall FB (2008) Checklist of nemertean genera and species published between 1995 and 2007. Spec Diver 13:245–274
Chen HX, Sundberg P, Norenburg JL, Sun SC (2009) The complete mitochondrial genome of Cephalothrix simula (Iwata) (Nemertea: Palaeonemertea). Gene 442:8–17. doi:10.1016/j.gene.2009.04.015
Podsiadlowski L, Braband A, Struck TH, von Döhren J, Bartolomaeus T (2009) Phylogeny and mitochondrial gene order variation in Lophotrochozoa in the light of new mitogenomic data from Nemertea. BMC Genomics 10:364. doi:10.1186/1471-2164-10-364
Turbeville JM, Smith DM (2007) The partial mitochondrial genome of the Cephalothrix rufifrons (Nemertea, Palaeonemertea): characterization and implications for the phylogenetic position of Nemertea. Mol Phylogenet Evol 43:1056–1065. doi:10.1016/j.ympev.2006.10.025
Chen HX, Strand M, Norenburg JL, Sun SC, Kajihara H, Chernyshev AV, Maslakova SA, Sundberg P (2010) Statistical parsimony networks and species assemblages in cephalotrichid nemerteans (Nemertea). PLoS ONE 5:e12885. doi:10.1371/journal.pone.0012885
Sambrook JRD (2001) Rapid isolation of yeast DNA. In: Sambrook JRD (ed) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, pp 631–632
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299
Palumbi S (1996) Nucleic acids: II. the polymerase chain reaction. In: Hillis DM, Moritz C, Mable BK (eds) Molecular systematics. Sinauer Associates, Sunderland, pp 205–247
Boore JL, Brown WM (2000) Mitochondrial genomes of Galathealinum, Helobdella, and Platynereis: sequence and gene arrangement comparisons indicate that Pogonophora is not a phylum and Annelida and Arthropoda are not sister taxa. Mol Biol Evol 17:87–106
Hall J (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599. doi:10.1093/molbev/msm092
Perna NT, Kocher TD (1995) Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol 41:353–358
Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
De Rijk P, De Wachter R (1997) RnaViz, a program for the visualisation of RNA secondary structure. Nucleic Acids Res 25:4679–4684
Stothard P, Wishart DS (2005) Circular genome visualization and exploration using CGView. Bioinformatics 21:537–539. doi:10.1093/bioinformatics/bti054
Serb JM, Lydeard C (2003) Complete mtDNA sequence of the north American freshwater mussel, Lampsilis ornata (Unionidae): an examination of the evolution and phylogenetic utility of mitochondrial genome organization in Bivalvia (Mollusca). Mol Biol Evol 20:1854–1866. doi:10.1093/molbev/msg218
Boore JL (2006) The complete sequence of the mitochondrial genome of Nautilus macromphalus (Mollusca: Cephalopoda). BMC Genomics 7:182. doi:10.1186/1471-2164-7-182
Yokobori SI, Pääbo S (1995) tRNA editing in metazoans. Nature 377:490. doi:10.1038/377490a0
Lavrov DV, Brown WM, Boore JL (2000) A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. Proc Natl Acad Sci USA 97:13738–13742. doi:10.1073/pnas.250402997
Masta SE, Boore JL (2004) The complete mitochondrial genome sequence of the spider Habronattus oregonensis reveals rearranged and extremely truncated tRNAs. Mol Biol Evol 21:893–902. doi:10.1093/molbev/msh096
Wolstenholme DR (1992) Animal mitochondrial DNA: structure and evolution. Int Rev Cytol 141:173–216
Anderson S, Bankier AT, Barrell BG et al (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465
Ojala D, Montoya J, Attardi G (1981) tRNA punctuation model of RNA processing in human mitochondria. Nature 290:470–474
Ojala D, Merkel C, Gelfand R, Attardi G (1980) The tRNA genes punctuate the reading of genetic information in human mitochondrial DNA. Cell 22:393–403. doi:10.1016/0092-8674(80)90350-5
Montoya J, Gaines GL, Attardi G (1983) The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell 34:151–159. doi:10.1016/0092-8674(83)90145-9
Boore JL, Brown WM (1994) Complete DNA sequence of the mitochondrial genome of the black chiton, Katharina tunicata. Genetics 138:423–443
Kim I, Lee EM, Seol KY, Yun EY, Lee YB, Hwang JS, Jin BR (2006) The mitochondrial genome of the Korean hairstreak, Coreana raphaelis (Lepidoptera: Lycaenidae). Insect Mol Biol 15:217–225. doi:10.1111/j.1365-2583.2006.00630.x
Fenn JD, Cameron SL, Whiting MF (2007) The complete mitochondrial genome sequence of the Mormon cricket (Anabrus simplex: Tettigoniidae: Orthoptera) and an analysis of control region variability. Insect Mol Biol 16:239–252. doi:10.1111/j.1365-2583.2006.00721.x
Jermiin LS, Graur D, Lowe RM, Crozier RH (1994) Analysis of directional mutation pressure and nucleotide content in mitochondrial cytochrome b genes. J Mol Evol 39:160–173
Francino MP, Ochman H (1997) Strand asymmetries in DNA evolution. Trends Genet 13:240–245. doi:10.1016/S0168-9525(97)01118-9
Hassanin A, Leger N, Deutsch J (2005) Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of metazoa, and consequences for phylogenetic inferences. Syst Biol 54:277–298
Clary DO, Wolstenholme DR (1985) The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol 22:252–271
Boore JL, Brown WM (1995) Complete sequence of the mitochondrial DNA of the annelid worm Lumbricus terrestris. Genetics 141:305–319
Ikemura T (1982) Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J Mol Biol 158:573–597. doi:10.1016/0022-2836(82)90250-9
Sharp PM, Matassi G (1994) Codon usage and genome evolution. Curr Opin Genet Dev 4:851–860
Duret L, Mouchiroud D (1999) Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc Natl Acad Sci USA 96:4482–4487
Helfenbein KG, Brown WM, Boore JL (2001) The complete mitochondrial genome of the articulate brachiopod Terebratalia transversa. Mol Biol Evol 18:1734–1744
Broughton RE, Reneau PC (2006) Spatial covariation of mutation and nonsynonymous substitution rates in vertebrate mitochondrial genomes. Mol Biol Evol 23:1516–1524. doi:10.1093/molbev/msl013
Yang Z, Bielawski JP (2000) Statistical methods for detecting molecular adaptation. Trends Ecol Evol 15:496–503
Hurst LD (2002) The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet 18:486–487
Li W (1997) Rates and patterns of nucleotide substitution. In: Andrew DS (ed) Molecular evolution. Sinauer Associates, Sunderland, pp 177–210
Macey JR, Larson A, Ananjeva NB, Papenfuss TJ (1997) Replication slippage may cause parallel evolution in the secondary structures of mitochondrial transfer RNAs. Mol Biol Evol 14:30–39
Yamazaki N, Ueshima R, Terrett JA et al (1997) Evolution of pulmonate gastropod mitochondrial genomes: comparisons of gene organizations of Euhadra, Cepaea and Albinaria and implications of unusual tRNA secondary structures. Genetics 145:749–758
Okimoto R, Macfarlane JL, Clary DO, Wolstenholme DR (1992) The mitochondrial genomes of two nematodes, Caenorhabditis elegans and Ascaris suum. Genetics 130:471–498
Yokobori S, Pääbo S (1997) Polyadenylation creates the discriminator nucleotide of chicken mitochondrial tRNA(Tyr). J Mol Biol 265:95–99
Jacobs HT, Elliott DJ, Math VB, Farquharson A (1988) Nucleotide sequence and gene organization of sea urchin mitochondrial DNA. J Mol Biol 202:185–217. doi:10.1016/0022-2836(88)90452-4
Boyce TM, Zwick ME, Aquadro CF (1989) Mitochondrial DNA in the bark weevils: size, structure and heteroplasmy. Genetics 123:825–836
L’Abbe D, Duhaime JF, Lang BF, Morais R (1991) The transcription of DNA in chicken mitochondria initiates from one major bidirectional promoter. J Biol Chem 266:10844–10850
Kolpakov R, Bana G, Kucherov G (2003) mreps: efficient and flexible detection of tandem repeats in DNA. Nucleic Acids Res 31:3672–3678
Curole JP, Kocher TD (1999) Mitogenomics: digging deeper with complete mitochondrial genomes. Trends Ecol Evol 14:394–398
Moritz C, Brown WM (1987) Tandem duplications in animal mitochondrial DNAs: variation in incidence and gene content among lizards. Proc Natl Acad Sci USA 84:7183–7187
Stanton DJ, Daehler LL, Moritz CC, Brown WM (1994) Sequences with the potential to form stem-and-loop structures are associated with coding-region duplications in animal mitochondrial DNA. Genetics 137:233–241
Saccone C, De Giorgi C, Gissi C, Pesole G, Reyes A (1999) Evolutionary genomics in Metazoa: the mitochondrial DNA as a model system. Gene 238:195–209. doi:10.1016/S0378-1119(99)00270-X
Vallès Y, Boore JL (2006) Lophotrochozoan mitochondrial genomes. Integr Comp Biol 46:544–557. doi:10.1093/Icb/Icj056
Shen X, Ma X, Ren J, Zhao F (2009) A close phylogenetic relationship between Sipuncula and Annelida evidenced from the complete mitochondrial genome sequence of Phascolosoma esculenta. BMC Genomics 10:136. doi:10.1186/1471-2164-10-136
Noguchi Y, Endo K, Tajima F, Ueshima R (2000) The mitochondrial genome of the brachiopod Laqueus rubellus. Genetics 155:245–259
Nakao M, Sako Y, Ito A (2003) The mitochondrial genome of the tapeworm Taenia solium: a finding of the abbreviated stop codon. U J Parasitol 89:633–635
Plaisance L, Huyse T, Littlewood DT, Bakke TA, Bachmann L (2007) The complete mitochondrial DNA sequence of the monogenean Gyrodactylus thymalli (Platyhelminthes: Monogenea), a parasite of grayling (Thymallus thymallus). Mol Biochem Parasitol 154:190–194. doi:10.1016/j.molbiopara.2007.04.012
Steinauer ML, Nickol BB, Broughton R, Orti G (2005) First sequenced mitochondrial genome from the phylum Acanthocephala (Leptorhynchoides thecatus) and its phylogenetic position within Metazoa. J Mol Evol 60:706–715. doi:10.1007/s00239-004-0159-8
Min GS, Park JK (2009) Eurotatorian paraphyly: revisiting phylogenetic relationships based on the complete mitochondrial genome sequence of Rotaria rotatoria (Bdelloidea: Rotifera: Syndermata). BMC Genomics 10:533. doi:10.1186/1471-2164-10-533
Fearnley IM, Walker JE (1986) Two overlapping genes in bovine mitochondrial DNA encode membrane components of ATP synthase. EMBO J 5:2003–2008
Dunn CW, Hejnol A, Matus DQ et al (2008) Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452:745–749. doi:10.1038/nature06614
Struck TH, Fisse F (2008) Phylogenetic position of Nemertea derived from phylogenomic data. Mol Biol Evol 25:728–736. doi:10.1093/molbev/msn019
Rawlings TA, Collins TM, Bieler R (2001) A major mitochondrial gene rearrangement among closely related species. Mol Biol Evol 18:1604–1609
Morrison CL, Harvey AW, Lavery S, Tieu K, Huang Y, Cunningham CW (2002) Mitochondrial gene rearrangements confirm the parallel evolution of the crab-like form. Proc R Soc Lond B 269:345–350. doi:10.1098/rspb.2001.1886
Lavrov DV, Brown WM, Boore JL (2004) Phylogenetic position of the Pentastomida and (pan)crustacean relationships. Proc R Soc Lond B 271:537–544. doi:10.1098/rspb.2003.2631
Akasaki T, Nikaido M, Tsuchiya K, Segawa S, Hasegawa M, Okada N (2006) Extensive mitochondrial gene arrangements in coleoid Cephalopoda and their phylogenetic implications. Mol Phylogenet Evol 38:648–658. doi:10.1016/j.ympev.2005.10.018
Faure E, Casanova JP (2006) Comparison of chaetognath mitochondrial genomes and phylogenetical implications. Mitochondrion 6:258–262. doi:10.1016/j.mito.2006.07.004
Acknowledgments
This work was supported by the National Natural Science Foundation of China (30970333) and the Swedish Research Council (80565801) (to PS). We are grateful to Wei Shi and Dong-Li Xu for assisting with the analyses.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, HX., Sundberg, P., Wu, HY. et al. The mitochondrial genomes of two nemerteans, Cephalothrix sp. (Nemertea: Palaeonemertea) and Paranemertes cf. peregrina (Nemertea: Hoplonemertea). Mol Biol Rep 38, 4509–4525 (2011). https://doi.org/10.1007/s11033-010-0582-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0582-4