Abstract
A molecular study was conducted to investigate the prevalence of Hepatitis C virus genotypes in HCV infected population of UAE. 67 HCV seropositive samples were collected from different health care centres. Quantitative analysis of these samples using PCR resulted in 67 positive samples. The PCR positive samples were subjected to genotyping using the method described by Simmonds et al. (J Gen Virol 74: 2391–2399, 1993). HCV genotype 4 was the predominant genotype (46.2%) followed by genotype 3a (23.8%) and 1a (15%). The predominant genotype among the female patients was genotype 4 (65.6%), while genotype 3a was the predominant among the male patients (42.8%). The predominance of HCV genotype 4 in our population confirms the predominance of HCV genotype 4 in UAE and most of the Arab countries in the Middle East. Implications of genotyping for clinical outcome of HCV infection, response to treatment as well as for vaccine development are discussed.
Similar content being viewed by others
Introduction
It is well established now that hepatitis C virus (HCV) is the leading cause of blood borne non-A, non-B hepatitis worldwide. Hepatitis is one of the major causes of morbidity and mortality developing countries including UAE [1–4].
Acute hepatitis C is mild and often asymptomatic while chronic hepatitis C is an indolent course but may progress to cirrhosis and HCC [5]. It is a slowly progressive infection spread primarily through intravenous drug users, can also spread by sharing of tooth brushes, razors and contaminated needles, sexual relations, from mother to child, etc. HCV RNA has not been detected in semen, urine, stool or vaginal secretion and weather it is present in saliva remains controversial [6]. An estimate of 53,000 deaths per year caused due to HCV in world. Most HCV infected people remained unidentified until the development of late symptoms, while some remained carrier through their life and do not develop any complication.
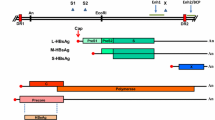
HCV identified in 1989 belongs to family Flaviviridae. Its whole genome has been sequenced and identified. The viral particle consists of an envelope derived from host membranes into which are inserted the virally encoded glycoproteins (E1 and E2) surrounding a nucleocapsid and a positive sense, single stranded RNA genome which has been identified and sequenced [5]. The whole genome of 9,500 nucleotides contains highly conserved untranslated regions (UTR) at both the 50 and 30 termini, which flank a large translational open reading frame encoding a poly-protein of 3,000 amino acids. This is processed by both cellular and viral proteases to produce the specific viral gene products. The structural proteins (core, E1 and E2) are located in the N-terminal quarter while non-structural (NS) proteins (NS2, NS3, NS4A, NS4B, NS5A, NS5B) in the remaining portion of the polyprotein. Early analyzes of the 30 UTR resulted in conflicting data regarding its exact sequence content. Most studies indicated that the genome terminated with a poly (U) tract, while one group reported a poly (A). These differences in the 30 UTR were considered unusual given the importance of these generally well-conserved untranslated regions in the RNA replication of other positive-stranded viruses. The 50 UTR is the most conserved portion of the HCV genome, although nucleotide variations characteristic of different HCV types exist that have been used in polymerase chain reaction based genotyping assays [7].
HCV has population specific genotype [8]. A recent proposal for HCV nomenclature defined six major genotypes [5–10] based upon phylogenetic analyzes of the core E1 and NS5 regions with further divisions into subtypes (1a, 1b, 2c, etc.). The genetic variability is due to high mutation rate in the envelope gene coupled with the absence of a proofreading function in the virion-encoded by RNA polymerase [8]. Researchers have not yet agreed whether or not five different viruses, three found only in Vietnamese individuals, the remaining two seen in Indonesians, are actually subtypes of genotype 6 or ought to be designated as genotypes 7 through 11. Some genotypes (1a, 1b, 2a, 2b, 3a) are widely distributed around the world [11, 12], while others have a more restricted distribution. Genotype 4 is predominant in the Middle East (particularly Egypt), Zaire and Burundi [13–15], while genotype 5 has so far been mainly found in South Africa [16]. There is increasing evidence that the HCV genotype with which an individual is infected may have important implications for the clinical outcome of infection and its response to interferon treatment [17–19]. Thus, typing of HCV isolates becomes an additional tool in the diagnosis and treatment of HCV infection [20].
HCV genotyping provides valuable epidemiological and therapeutic information. Current therapy, which consists of a combination of pegylated interferon and ribavirin, gives a response rate of between 48 (genotypes 1, 4, 5 and 6) and 88% for genotypes 2 and 3 [9]. The duration of therapy is variable as well for different genotypes. These findings indicate the importance of genotype knowledge before therapy. In this connection our studies can be very valuable for the health care providers and clinicians in designing the therapeutic strategies to cope this manic disease in this part of the world. As such kind of study has not yet been performed in this area.
Material and methods
Ethics statement
The study involved 67 UAE patients who were positive for anti-HCV by ELISA. A blood sample was collected during 2006–2007 from each of these patients during one of their regular follow-up visits. Written consent was not taken because all the investigations done in this research were routinely for all hepatitis C patients including the HCV genotyping, beside no patients identification (name or number) where mentioned in this research.
Sera were separated from the samples and stored at −70 C, in 200 μl aliquots, until further processing.
Testing for anti-HCV was repeated in all anti-HCV positive patients using a standard ELISA (bioMerieux); and, if both of these assays gave a positive result, a line immunoassay (INNO-LIA HCV Ab III; Innogenetics, Ghent, Belgium) was performed. Only samples found positive in the line immunoassay were considered positive for anti-HCV. Detection of HCV RNA was attempted on a 500 μl sample of each serum found positive for anti-HCV, using a commercial, PCR-based test (Taqman amplicor, Roche). The manufacturere’s in structions were followed and the internal control supplied by the manufacturer was added to each specimen, as an extraction and amplification control.
HCV genotyping was performed using hybridization with sequence-specific oligonucleotides and as described before [21].
The χ2 test was used to determine the variation in genotype distribution between the different groups. Statistical significance was considered as P < 0.05. All statistical tests were performed using SPSS statistical software package.
Results
HCV genotype distribution in 67 HCV-positive UAE patients is shown in Table 1. Overall, HCV genotype 4 was the predominant genotype (46.2%) followed by genotype 3a (23.8%) and 1a (15%). The predominant genotype among the female patients was genotype 4 (65.6%), while genotype 3a was the predominant among the male patients (42.8%). Differences in genotype distribution were statistically significant.
HCV genotype distribution among HCV-positive UAE patients is similar to that in other Middle Eastern Arab countries except for Jordan where genotype 1a predominates (Table 2). In Middle Eastern non-Arab countries (Iran, and Turkey) genotype 4 was minimally detected and genotypes 1a or 1b were the predominant genotypes.
Discussion
HCV is known to have marked genetic heterogeneity with nucleotide substitution rate of 1.44 9 10−3 and 1.92 9 10−3 per site per year [10, 22]. Accumulation of nucleotide substitutions in the HCV genome results in diversification and evolution into different genotypes. Presently, HCV can be classified into at least six major and a series of subtypes [13, 23]. There is increasing evidence that patients infected with different genotypes may have different clinical profiles, severity of liver diseases, and response to alpha interferon therapy [24–28]. Hence, a convenient and reliable HCV genotyping system is essential for large epidemiological and clinical studies. In this context, a genotyping method, based on genotype specific primers for PCR of the core gene, by which HCV isolates can be classified into genotypes was described [8].
To our knowledge, this is the first published study to report on the genotyping of HCV in different groups of HCV-positive patients in UAE, representing close to a population-based sample. Our results show that HCV genotype 4 is the predominant genotype (46.2%) followed by 3a (23.8%) and 1a (15%). HCV genotype 4 is the predominant genotype among females (65.6%). HCV genotype 3a is the predominant genotype among males (42.8%).
This difference in genotype distribution can perhaps be explained in light of the finding that genotype influences clinical outcome [29]. Zekri et al. [29] showed that infection with genotypes 1a and 4 may be considered a risk factor for the induction of neu-oncoprotein overexpression and subsequent development of hepatocellular carcinoma (HCC).
The predominance of HCV genotype 4 in the UAE population (46.2%) is in agreement with other reports on genotyping of HCV isolates in different Middle Eastern countries. Table 2 summarizes only the studies with relatively large numbers investigated [29–34]. It is of interest to note that in contrast to Arab countries genotype 4 can hardly be detected in non-Arab Middle Eastern countries such as Iran [35], and Turkey [36]. The presence of other genotypes such as 2a, and 1b among UAE patients can be attributed to many factors. These include the expatriates from deferent nationalities resided in UAE for quite some time and participated in blood donation.
According to the World Health Organization, 180 million individuals in the world are infected with HCV and this is a growing global problem. The development of an effective vaccine remains the ideal way to combat HCV infection. In addition to the implications for clinical outcome of infection, and for treatment, genotyping of HCV also has major implications for HCV vaccine development. Recent data suggest that for a vaccine to be fully protective it should contain a range of deferent envelope proteins corresponding to the common genotypes in particular geographic regions. Vaccines for use in the Middle East should, therefore, not be based only on genotype 4 sequences; other genotypes such as 1a and 1b are also equally important. Finally, genotyping of HCV may be a useful epidemiological marker particularly in establishing suspected unconventional routes of HCV transmission such as vertical [37], intraspousal or interfamilial transmission [38].
References
Millor J, Holmes EC, Jarvis LM, Yap PL, Simmonds P (1995) The international HCV collaborative study group. Investigation of the pattern of hepatitis C virus sequence diversity in different geographical regions: implications for virus classification. J Gen Virol 76:2493–2507
Simmonds P, Mellor J, Sakuldamrongpanich T, Nuchaprayoon C, Tanprasert S, Holmes EC, Smith DB (1996) Evolutionary analysis of variants of hepatitis C virus found in Southeast Asia: comparison with classifications based upon sequence similarity. J Gen Virol 77:3013–3024
de Lamballerie X, Charrel RN, Attoui H, De Micco P (1997) Classification of hepatitis C virus variants in six major types based on analysis of the envelop 1 and non-structural 5B genome regions and complete polyprotein sequences. J Gen Virol 78:45–51
Alter MJ (1995) Epidemiology of hepatitis V in the West. Semin Liver Dis 15:5–14
Bhupinder N, Bhandari MD, Teresa L, Wright MD (1995) Hepatitis C: an overview. Annu Rev Med 46:309–317. doi:10.1146/annurev.med.46.1.309
Hsu H, Wright TL, Luba D (1991) Failure to detect hepatitis C virus genome in human secretion with PCR. Hepatology 14:753–767. doi:10.1002/hep.1840140504
Major ME, Feinstone SM (1997) The molecular virology of Hepatitis C. Hepatology 25:1527–1538. doi:10.1002/hep.510250637
Ohno T, Mezokami M, Wu R, Saleh MG, Ohba K, Orito E et al (1997) New Hepatitis C Virus (HCV) genotyping system that allows for identification of HCV Genotypes 1a, 1b, 2a, 2b, 3a, 3b, 4, 5a, and 6a. J Clin Microbiol 35:201–202
Poynard T, McHutchion JG, Goodman ZD, Ling MH, Albretch J (2000) Is an ‘‘A la carte’’ combination interferon alpha-2b plus ribavirin regimen possible for the first line treatment in patients with chronic Hepatitis C? ALGOVIRC Proj group 31:211–218
Ogata N, Alter HJ, Miller RH, Purcel RH (1991) Nucleotide sequence and mutation rate of the H strain of Hepatitis C virus. Proc Natl Acad Sci USA 88(5):3392–3396
McOmish F, Yap PL, Dow BC, Follett EA, Seed C, Keller AJ, Cobain TJ, Krusius T, Kolho E, Naukkarinen R (1994) Geographical distribution of hepatitis C virus genotypes in blood donors–an inter- national collaborative survey. J Clin Microbiol 32:884–892
Schreier E, Roggendorf M, Driesel G, Höhne M, Viazov S (1996) Genotypes of hepatitis C virus isolates from different parts of the world. Arch Virol (suppl)11:185–193
Bukh J, Purcell RH, Miller RH (1994) Sequence analysis of core gene of 14 hepatitis C virus genotypes. Proc Natl Acad Sci USA 91:8239–8243
Simmonds P, McOmish F, Yap PL, Chan SW, Lin CK, Dusheiko G, Saeed AA (1993) Holmes EC sequence variability in the 5 k non coding region of hepatitis C virus: identification of a new virus type and restriction on sequence diversity. J Gen Virol 74:661–668
Stuyver L, van Arnhem W, Wyseur A, Hernandez F, Delaporte E, Maertens G (1994) Classification of hepatitis C Viruses based on phylogenetic analysis of the envelope E1 and non-structural 5B regions and identification of five additional subtypes. Proc Natl Acad Sci USA 91:10134–10138
Davidson F, Simmonds P, Ferguson JC, Jarvis LM, Dow BC, Follett EA, Seed CR, Krusius T, Lin C, Medgyesi GA (1995) Survey of major genotypes and subtypes of hepatitis C virus using RFLP of sequences amplified from the 5 k non-coding region. J Gen Virol 76:1197–1204
Mondelli MU, Silini E (1999) Clinical significance of hepatitis C virus genotypes. J Hepatol 31:65–70
al-Faleh FZ, Sbeih F, al-Karawi M, al-Mofleh IA, al-Rashed RS, Ayoola A, al-Amri S, Mayet I, al-Habbal TM, al-Omair A, al-Sohaibani MO, Abdullah AO, Mohamed SA, el-Sheikh MA (1998) Treatment of chronic hepatitis C genotype 4 with alpha-interferon in Saudi Arabia: a multicenter study. Hepato-gastroenterology 45:488–491
Zein NN (2000) Clinical significance of hepatitis C virus genotypes. Clin Microbiol Rev 13:223–235
Roggendorf M, Lu M, Meisel H, Riffelmann M, Schreier E, Viazov S (1996) Rational use of diagnostic tools in hepatitis C. J Hepatol 24:26–34
Simmonds P, Holmes EC, Cha TA, Chan SW, McOmish F, Irvine B, Beall E, Yap PL, Kolberg J, Urdea MS (1993) Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol 74(Pt 11):2391–2399
Okamoto H, Kurai K, Okada S, Yamammoto K, Lizuka H, Tanaka T et al (1992) Full-length sequence of a Hepatitis C virus genome having poor homology to reported isolates: comparatively study of four distinct genotypes. Virology 188:331–341. doi:10.1016/0042-6822(92)90762-E
Ohba K, Mizokami M, Ohno T, Suzuki K, Orito E, Ina Y et al (1995) Classification of Hepatitis C virus into major types and subtypes based on molecular phylogenic tree analysis. Virus Res 36:201–214. doi:10.1016/0168-1702(95)00007-D
Kanai K, Kako M, Okamoto H (1992) HCV genotypes in chronic hepatitis C virus and response to interferon. Lancet 339:1543. doi:10.1016/0140-6736(92)91311-U
Mita E, Hayashi N, Kanazawa Y, Hagiwara H, Ueda K, Kasahara A et al (1994) Hepatitis C virus genotype and RNA titer in the progression of type C chronic liver disease. J Hepatol 21:468–473. doi:10.1016/S0168-8278(05)80330-7
Pozzato G, Moretti M, Franzin F, Croce LS, Tribelli C, Masayu T et al (1991) Severity of liver disease with different Hepatitis C viral clones. Lancet 338:509. doi:10.1016/0140-6736(91)90578-D
Saito I, Miyyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe Y, Koi S, Onji M, Ohta Y, Choo LQ, Houghton M, Kuo G (1990) Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA 87:6547–6549. doi:10.1073/pnas.87.17.6547
Yoshioka K, Kakumu S, Wakita T, Ishikawa T, Itoh Y, Takayanagi M et al (1992) Detection of hepatitis C virus by polymerase chain reaction and response to interferon alpha therapy: relationship to genotypes of Hepatitis C virus. Hepatology 16:293–299. doi:10.1002/hep.1840160203
Zekri AR, Bahnassy AA, Shaarawy SM, Mansour OA, Maduar MA, Khaled HM, El-Ahmadi O (2000) Hepatitis C virus genotyping in relation to neu-oncoprotein overexpression and the development of hepatocellular carcinoma. J Med Microbiol 49:89–95
Zein NN, Rakela J, Krawitt EL, Reddy KR, Tominaga T, Persing DH (1996) Hepatitis C virus genotypes in the United States: epidemiology, pathogenicity, and response to interferon therapy. Ann Intern Med 125:634–639
Bdour S (2002) Hepatitis C virus infection in Jordanian hemodialysis units: serological diagnosis and genotyping. J Med Microbiol 51:200–204
Al-Ahdal M, Rezeig M, Kessie G (1997) Genotyping of hepatitis C virus isolates from Saudi patients by analysis of sequences from PCR-amplified core region of the virus genome. Ann Saudi Med 17:601–604
Shobokshi OA, Serebour FE, Skakni LL (2003) Hepatitis C genotypes/subtypes among chronic hepatitis patients in Saudi Arabia. Saudi Med J 24(Suppl. 2):S87–S91
Abdulkarim AS (1996) Hepatitis C virus genotypes and polymorphism analysis of 5 k noncoding region. J Clin Microbiol 34:2623–2624
Sawant P (1998) Genotypes of GB virus C and hepatitis C virus in hepatitis patients in India, Iran and Slovenia. Hepatol Res 10:175–183
Bozdayi G, Rota S, Verdi H, Derici U, Sindel S, Bali M, Başay T (2002) The presence of hepatitis C virus infection in hemodialysis patients and determination of HCV genotype distribution. Mikrobiyol Bulletine 36:291–300
Mastromatteo AM, Rapaccini GL, Pompili M, Ursino S, Romano-Spica V, Gasbarrini G, Vanini G (2001) Hepatitis C virus infection: other biological fluids than blood may be responsible for intrafamilial spread. Hepato-gastroenterology 48:193–196
Sharara AI, Ramia S, Ramlawi F, Fares JE, Klayme S, Naman R (2007) Genotypes of hepatitis C virus (HCV) among positive Lebanese patients: comparison of data with that from other Middle Eastern countries. Epidemiol Infect 135(3):427–432
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Alfaresi, M.S. Prevalence of hepatitis C virus (HCV) genotypes among positive UAE patients. Mol Biol Rep 38, 2719–2722 (2011). https://doi.org/10.1007/s11033-010-0415-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0415-5