Abstract
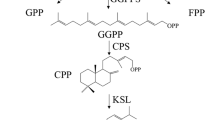
Salvia miltiorrhiza is a valuable Chinese herb (Danshen) that is widely used in traditional Chinese medicine. Diterpene quinones, known as tanshinones, are the main bioactive components of S. miltiorrhiza; however, there is only limited information regarding the molecular mechanisms underlying secondary metabolism in this plant. We used cDNA microarray analysis to identify changes in the gene expression profile at different stages of hairy root development in S. miltiorrhiza. A total of 203 genes were singled out from 4,354 cDNA clones on the microarray, and 114 unique differentially expressed cDNA clones were identified: six genes differentially expressed in 45-day hairy root compared with 30-day hairy root; 96 genes differentially expressed in 60-day hairy root compared with 30-day hairy root; and 12 genes unstably expressed at different stages. Among the 96 genes differentially expressed in 60-day hairy root compared with 30-day hairy root, a total of 57 genes were up-regulated, and 26 genes represent 29 metabolism-related enzymes. Copalyl diphosphate synthase, which catalyzes the conversion of the universal diterpenoid precursor (E,E,E)-geranylgeranyl diphosphate to copalyl diphosphate, was up-regulated 6.63 fold, and another six genes involved in tanshinone biosynthesis and eight candidate P450 genes were also differentially expressed. These data provide new insights for further identification of the enzymes involved in tanshinone biosynthesis.

Similar content being viewed by others
References
Wang XH, Morris-Natschke SL, Lee KH (2007) New developments in the chemistry and biology of the bioactive constituents of Tanshen. Med Res Rev 27(1):133–148
Hu ZB, Alfermann AW (1993) Diterpenoid production in hairy root cultures of S. miltiorrhiza. Phytochemistry 32(3):699–703
Shi M, Kwok KW, Wu JY (2007) Enhancement of tanshinone production in S. miltiorrhiza Bunge (red or Chinese sage) hairy-root culture by hyperosmotic stress and yeast elicitor. Biotechnol Appl Biochem 46(4):191–196
Gao W, Hillwig ML, Huang LQ, Cui GH, Wang XY, Kong JQ, Yang B, Peters RJ (2009) A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org Lett 11(22):5170–5173
Ohlrogge J, Benning C (2000) Unraveling plant metabolism by EST analysis. Curr Opin Plant Biol 3:224–228
Aharoni A, Vorst O (2002) DNA microarrays for functional plant genomics. Plant Mol Biol 48:99–118
Schena M, Shalon D, Davis RW, Brown PO (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270(5235):467–470
Cui GH, Huang LQ, Tang XJ, Qiu DY, Wang XY, Fu GF (2007) Functional genomics studies of Salvia miltiorrhizaI establish cDNA microarray of Salvia miltiorrhiza. China J Chin Mat Med 32(12):1137–1141
Shi YH, Zhu SW, Mao XZ, Feng JX, Qin YM, Zhang L, Cheng J, Wei LP, Wang ZY, Zhu YX (2006) Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18(3):651–664
Hihara Y, Kamei A, Kanehisa M, Kaplan A, Ikeuchi M (2001) DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13(4):793–806
The Gene Ontology Consortium (2000) Gene ontology: tool for the unification of biology. Nat Genet 25(1):25–29
Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M (2006) From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34:D354–D357
Cui GH, Huang LQ, Qiu DY, Yuan Y, Fu GF (2007) Functional genomics studies of Salvia miltiorrhiza II: gene expression profiling of different stage of hairy root. China J Chin Mater Med 32(13):1267–1272
Tian G, Zhang Y, Zhang T, Yang F, Ito Y (2000) Separation of tanshinones from Salvia miltiorrhiza Bunge by high-speed counter-current chromatography using stepwise elution. J Chromatogr A 904:107–111
Paseshnichenko VA (1998) A new alternative non-mevalonate pathway for isoprenoid biosynthesis in eubacteria and plants. Biochemistry (Mosc) 63:139–148
Hunter WN (2007) The non-mevalonate pathway of isoprenoid precursor biosynthesis. J Biol Chem 282:21573–21577
Eisenreich W, Bacher A, Arigoni D, Rohdich F (2004) Biosynthesis of isoprenoids via the non-mevalonate pathway. Cell Mol Life Sci 61:1401–1426
Buhaescu I, Izzedine H (2007) Mevalonate pathway: a review of clinical and therapeutical implications. Clin Biochem 40:575–584
Kuzuyama T, Seto H (2003) Diversity of the biosynthesis of the isoprene units. Nat Prod Rep 20:171–183
Kuzuyama T (2002) Mevalonate and nonmevalonate pathways for the biosynthesis of isoprene units. Biosci Biotechnol Biochem 66:1619–1627
Laule O, Furholz A, Chang HS, Zhu T, Wang X, Heifetz PB, Gruissem W, Lange M (2003) Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 100:6866–6871
Saito K, Hirai MY, Yonekura-Sakakibara K (2008) Decoding genes with coexpression networks and metabolomics–“Majority report by precogs”. Trends Plant Sci 13:36–43
Shulaev V, Cortes D, Miller G, Mittler R (2008) Metabolomics for plant stress response. Physiol Plant 132:199–208
Naoumkina MA, Modolo LV, Huhman DV (2010) Genomic and coexpression analyses predict multiple genes involved in triterpene saponin biosynthesis in Medicago truncatula. Plant Cell 22:850–866
Nelson DR, Schuler MA, Paquette SM, Werck-Reichhart D, Bak S (2004) Comparative genomics of rice and arabidopsis. Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiol 135:756–772
Schuler MA (1996) The role of cytochrome P450 monooxygenases in plant-insect interactions. Plant Physiol 112:1411–1419
Swaminathan S, Morrone D, Wang Q, Fulton DB, Peters RJ (2009) CYP76M7 is an ent-cassadine C11a-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell 21:3315–3325
Acknowledgments
We thank Dr. Deyou Qiu (Chinese academy of Forestry) for his helpful suggestion and Shanghai Biochip Company and National Engineering Research Center for Beijing Biochip Technology for microarray production and hybridization. This work was supported by grants from the Key Project of Chinese National Programs for Fundamental Research and Development (2006CB-504700), Chinese National Programs for High Technology Research and Development (2007AA02Z104) and independent studies supported by the Basic Scientific Research Expenses in China Academy of Chinese Medical Sciences (ZZ20090301).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cui, G., Huang, L., Tang, X. et al. Candidate genes involved in tanshinone biosynthesis in hairy roots of Salvia miltiorrhiza revealed by cDNA microarray. Mol Biol Rep 38, 2471–2478 (2011). https://doi.org/10.1007/s11033-010-0383-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0383-9