Abstract
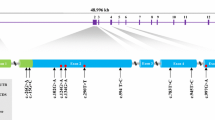
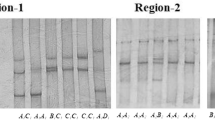
The cardiomyopathy associated 5 (CMYA5) gene was also called TRIM76, which was belonged to the tripartite motif super family of proteins (TRIM). It was a direct transcriptional target for MEF2A and it played an important role in myofibrillogenesis. In the present study, a 12056 bp cDNA sequence of the porcine CMYA5 gene was obtained by RT-PCR. The sequence encoded a large protein consisting of 4003 amino acids and the carboxyl terminus of the predicted CMYA5 protein comprised of a B-box coiled-coil, two fibronectin type III (FN3) repeats, and SPRY domains. The porcine CMYA5 gene was assigned to chromosome 2q21–24 by using the radiation hybrid (IMpRH) panel, and it was significantly linked to microsatellite Sw1602 with LOD scores of 6.74. Semi-quantitative RT–PCR revealed that the porcine CMYA5 gene was broadly expressed in all seven tissues(heart, liver, spleen, lung, kidney, skeletal muscle and adipose)harvested from different developmental stages(new born, five weeks and adult tongcheng pigs), with a high level in heart and skeletal muscle. One SNP (A7189C), leading to the amino acid alteration from the Ile residue to the Leu residue, was found and detected by BspTI PCR-restriction fragment length polymorphism. The association analysis revealed that the substitution of A7189C had significant associations with the percentage of ham (p < 0.05), water loss (p < 0.01) and intramuscular fat (p < 0.05). These results provide the evidence that the porcine CMYA5 gene can act as a potential candidate gene affecting pig meat quality.



Similar content being viewed by others
References
Tkatchenko AV, Piétu G, Cros N, Gannoun-Zaki L, Auffray C, Léger JJ, Dechesne CA (2001) Identification of altered gene expression in skeletal muscles from Duchenne muscular dystrophy patients. Neuromuscul Disord 11(3):269–277
Benson MA, Tinsley CL, Blake DJ (2004) Myospryn is a novel binding partner for dysbindin in muscle. J Biol Chem 279(11):10450–10458
Durham JT, Brand OM, Arnold M, Reynolds JG, Muthukumar L, Weiler H, Richardson JA, Naya FJ (2006) Myospryn is a direct transcriptional target for MEF2A that encodes a striated muscle, alpha-actinin-interacting, costamere-localized protein. J Biol Chem 281(10):6841–6849
Reynolds JG, McCalmon SA, Tomczyk T, Naya FJ (2007) Identification and mapping of protein kinase A binding sites in the costameric protein myospryn. Biochim Biophys Acta 1773(6):891–902
Kouloumenta A, Mavroidis M, Capetanaki Y (2007) Proper perinuclear localization of the TRIM-like protein myospryn requires its binding partner desmin. J Biol Chem 282(48):35211–35221
Reynolds JG, McCalmon SA, Donaghey JA, Naya FJ (2008) Deregulated protein kinase A signalling and myospryn expression in muscular dystrophy. J Biol Chem 283(13):8070–8074
Yerle M, Pinton P, Robic A, Alfonso A, Palvadeau Y, Delcros C, Hawken R, Alexander L, Beattie C, Schook L, Milan D, Gellin J (1998) Construction of a whole-genome radiation hybrid panel for high-resolution gene mapping in pigs. Cytogenet Cell Genet 82(3–4):182–188
Milan D, Hawken R, Cabau C, Leroux S, Genet C, Lahbib Y, Tosser G, Robic A, Hatey F, Alexander L, Beattie C, Schook L, Yerle M, Gellin J (2000) IMpRH server: an RH mapping server available on the web. Bioinformatics 16:558–559
Xu XL, Li K, Peng ZZ, Zhao SH, Yu M, Fan B, Zhu MJ, Xu SP, Du YQ, Liu B (2008) Molecular characterization, expression and association analysis with carcass traits of the porcine CMYA4 gene. J Anim Breed Genet 125:234–239
He X, Gao H, Liu C, Fan B, Liu B (2010) Cloning, chromosomal localization, expression profile and association analysis of the porcine WNT10B gene with backfat thickness. Mol Biol Rep. doi:10.1007/s11033-010-9978-4
Xu X, Qiu H, Du ZQ, Fan B, Rothschild MF, Yuan F, Liu B (2010) Porcine CSRP3: polymorphism and association analyses with meat quality traits and comparative analyses with CSRP1 and CSRP2. Mol Biol Rep 37(1):451–459
Liu K, Wang G, Zhao SH, Liu B, Huang JN, Bai X, Yu M (2009) Molecular characterization, chromosomal location, alternative splicing and polymorphism of porcine GFAT1 gene. Mol Biol Rep. 37:2711–2717
Nisole S, Stoye JP, Saïb A (2005) TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol 3(10):799–808
Torok M, Etkin LD (2001) Two B or not two B? Overview of the rapidly expanding B-box family of proteins. Differentiation 67(3):63–71
Goureau A, Yerle M, Schmitz A (1996) Human and porcine correspondence of chromosome segments using bidirectional chromosome painting. Genomics 36(2):252–262
Sun HF, Ernst CW, Yerle M, Pinton P, Rothschild MF, Chardon P, Rogel-Gaillard C, Tuggle CK (1999) Human chromosome 3 and pig chromosome 13 show complete synteny conservation but extensive gene-order differences. Cytogenet Cell Genet 85:273–278
Nezer C, Moreau L, Brouwers B, Coppieters W, Detilleux J, Hanset R, Karim L, Kvasz A, Leroy P, Georges M (1999) An imprinted QTL with major effect on muscle mass and fat deposition maps to the IGF2 locus in pigs. Nat Genet 21(2):155–156
Qu YC, Deng CY, Xiong YZ, Zheng R, Yu L, Su YH, Liu GL (2002) The construction of the genetic map and QTL locating analysis on chromosome 2 in swine. Yi Chuan Xue Bao 29(11):972–976
Kim JJ, Zhao H, Thomsen H, Rothschild MF, Dekkers JC (2005) Combined line-cross and half-sib QTL analysis of crosses between outbred lines. Genet Res 85(3):235–248
Lee SS, Chen Y, Moran C, Cepica S, Reiner G, Bartenschlager H, Moser G, Geldermann H (2003) Linkage and QTL mapping for Sus scrofa chromosome 2. J Anim Breed Genet 120(1):11–19
Malek M, Dekkers JC, Lee HK, Baas TJ, Prusa K, Huff-Lonergan E, Rothschild MF (2001) A molecular genome scan analysis to identify chromosomal regions influencing economic traits in the pig II. Meat and muscle composition. Mamm Genome 12(8):637–645
Rohrer GA, Thallman RM, Shackelford S, Wheeler T, Koohmaraie M (2005) A genome scan for loci affecting pork quality in a Duroc-Landrace F2 population. Anim Genet 37(1):17–27
Acknowledgements
We thank Dr Martine Yerle of INRA, France, for providing the RH panel. This research was supported by National Natural Science Foundation of China (30771536) and National High Science and Technology Foundation of China (2007AA10Z168).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xu, X., Xu, X., Yin, Q. et al. The molecular characterization and associations of porcine cardiomyopathy asssociated 5 (CMYA5) gene with carcass trait and meat quality. Mol Biol Rep 38, 2085–2090 (2011). https://doi.org/10.1007/s11033-010-0334-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0334-5