Abstract
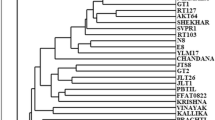
Finger millet (Eleusine coracana L.) is an important crop used for food, forage, and industrial products. Three DNA marker techniques, random amplified polymorphic DNA (RAPD), simple sequence repeat (SSR) and cytochrome P450 gene based markers were used for the detection of genetic polymorphism in 83 accessions of finger millet collected from various geographical regions of India and Africa. A total of 18 RAPD, 10 SSR and 10 pairs of cytochrome P450 gene based markers were generated 56.17, 70.19 and 54.29% polymorphism, respectively. Mean polymorphism information content (PIC) for each of these marker systems (0.280 for RAPD, 0.89 for SSR and 0.327 for cytochrome P450 gene based markers) suggested that SSR marker were highly effective in determining polymorphism. The phenograms based on the three markers data indicate that genotypes from different geographical regions are clearly distinguishable as separate clusters. Mantel test employed for detection of goodness of fit established cophenetic correlation values above 0.90 for all the three marker systems. The dendrograms and PCA plots derived from the binary data matrices of the three marker systems are highly concordant. High bootstrap values were obtained at major nodes of phenograms through WINBOOT software. Based on the results of present study, SSR and cytochrome P450 gene based markers appear to be particularly useful for the estimation of genetic diversity. This study reveals the potential of RAPD, SSR and gene based markers for characterizing germplasm of Eleusine coracana and narrow down the vast germplasm into distinct core groups.


Similar content being viewed by others
Abbreviations
- PCA:
-
Principal component analysis
- PCR:
-
Polymerase chain reaction
- PIC:
-
Polymorphism information content
- RAPD:
-
Random amplified polymorphic DNA
- UPGMA:
-
Unweighted pair group method with arithmetic averages
References
Venkannababu B, Ramana T, Radhakrisnan TM (1987) Chemical composition and protein content in hybrid varieties of finger millet. Indian J Agric Sci 57:520–522
Chennaveeraiah MS, Hiremath SC (1991) Cytogenetics of minor millets. In: Tsuchiya T, Gupta PK (eds) Chromosome engineering in plants genetics breeding and evolution. Elsevier, Amsterdam, pp 613–627
Chethan S, Malleshi NG (2007) Finger millet polyphenols: characterization and their nutraceutical potential. Am J Food Technol 27:282–292
Williams JGK, Kubelik AR, Livak KJ, Rafalski AJA, Tingey SV (1990) DNA polymorphism amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 22:6531–6535
Jacobson A, Hedrén M (2007) Phylogenetic relationships in Alisma Alismataceae based on RAPDs, and sequence data from ITS and trnL. Plant Syst Evol 265:27–44
Soller M, Beckmann JS (1983) Genetic polymorphism in varietal identification and genetic improvement. Theor Appl Genet 67:25–33
Vos PR, Hogers R, Bleeker M, Reijans M, Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for fingerprinting. Nucleic Acids Res 23(21):4407–4414
Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat SSR-anchored polymerase chain reaction amplification. Genomics 202:176–183
Becker J, Heun M (1994) Barley microsatellites: allele variation and mapping. Plant Mol Biol 274:835–845
Garcia AAF, Benchimol LL, Barbosa AMM, Geraldi IO, Souza CLJ, Souza AP (2004) Comparison of RAPD, RFLP, AFLP and SSR markers for diversity studies in tropical maize inbred lines. Genet Mol Biol 27:579–588
Nazari L, Pakniyat H (2008) Genetic diversity of wild and cultivated Barley genotypes under drought stress using RAPD markers. Biotechnology 74:745–750
Malik AR, Pervaiz ZH, Masood MS (2008) Genetic diversity analysis of traditional and improved cultivars of Pakistani rice Oryza sativa L. using RAPD markers. Electron J Biotechnol 11(3):1–10
Davierwal AAP, Chowdari KV, Shiv K, Reddy APK, Ranjekar PK, Gupta VS (2000) Use of three different marker systems to estimate genetic diversity of Indian elite rice varieties. Genetica 108:269–284
Rashed MA, Abou-Deif MH, Sallam MAA, Ramadan WA (2008) Estimation of genetic diversity among thirty bread wheat varieties by RAPD analysis. J Appl Sci Res 412:1898–1905
Colombo C, Second G, Valle TL, Charrier A (1998) Genetic diversity characterization of cassava cultivars Manihot esculenta Crantz. I. RAPD markers. Genet Mol Biol 21:69–84
Chao S, Zhang W, Dubcovsky J, Sorrells M (2007) Evaluation of genetic diversity and genome wide linkage is equilibrium among US wheat triticum aestivum L germplasm representing different market classes. Crop Sci 47:1018–1030
Agrama HA, Tuinstra MR (2003) Phylogenetic diversity and relationships among sorghum accessions using SSRs and RAPDs. Afr J Biotechnol 2:334–340
Qi-lun Y, Ping F, Ke-cheng K, Guang-Tang P (2008) Genetic diversity based on SSR markers in maize Zea mays L. landraces from Wuling mountain region in China. J Genet 87:287–291
Shalk M, Nedelkina S, Schoch G, Batard Y, Werck-reichhart D (1999) Role of unusual amino-acid residues in the proximal and distal heme regions of a plant P450, CYP73A1. Biochemistry 38:6093–6103
Tanksley SD, Mccouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Somerville C, Somerville S (1999) Plant functional genomics. Science 285:380–383
Yamanaka S, Suzuki E, Tanaka M, Takeda Y, Watanabe JA, Watanabe KN (2003) Assessment of cytochrome P450 sequences offers a useful tool for determining genetic diversity in higher plant species. Theor Appl Genet 108:1–9
Salimath SS, Olivera ACD, Godwin ID, Bennetzen JL (1995) Assessment of genome origins and diversity in the genus Eleusine with DNA markers. Genome 38:757–763
Fakrudin B, Kulkarni RS, Shashidhar HE, Hittalmani S (2007) Genetic diversity assessment of finger millet, Eleusine coracana, germplasm through RAPD analysis. PGR Newslett 138:52–54
Babu B, Senthil N, Gomez S, Biji K, Rajendraprasad N, Kumar S, Babu R (2007) Assessment of genetic diversity among finger millet Eleusine coracana L Gaertn accessions using molecular markers. Genet Resour Crop Evol 54:399–404
Parani M, Rajesh K, Lakshmi M, Parducci L, Szmidt AE, Parida A (2001) Species identification in seven small milletspecies using polymerase chain reaction–restriction fragment length polymorphism oftrnS-psbC gene region. Genome 44:495–499
Hilu KW (1995) Evolution of finger millet, evidence from random amplified polymorphic DNA. Genome 38:232–238
Dida MM, Wanyera N, Dunn MLH, Bennetzen JL, Devos KM (2008) Population structure and diversity in finger millet Eleusine coracana germplasm. Trop Plant Biol 1:131–141
Dida MM, Srinivasachary, Ramakrishnan S, Bennetzen JL, Gale MD, Devos KM (2007) The genetic map of finger millet, Eleusine coracana. Theor Appl Genet 114:321–332
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4326
Maniatis T, Sambrook J, Fritsch EF (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Rohlf FJ (2001) NTSYS-pc numerical taxonomy and multivariate analysis system. Version 5.1. Exeter Publishing Ltd., Setauket
Jaccard P (1908) Nouvelles recherches sur la distribuition florale. Bull Soc Vaud Sci Nat 44:223–270
Mental N (1967) The detection of disease clustering and generalized regression approach. Cancer Res 27:209–220
Yap IV, Nelson R (1995) WinBoot: a program for performing bootstrap analysis of binary data to determine the confidence limits of UPGMA-based dendrograms. IRRI discussion paper series no. 14. IRRI, Los Banos
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR microsatellite marker for germplasm analysis. Mol Breed 2:225–238
Cooke RJ (1995) Variety identification of crop plants. In: Skerrit JH, Appels R (eds) New diagnostics in crop science. Biotechnology in Agriculture No. 13 CAB International, Wallingford, UK, pp 33–63
Vadivoo AS, Joseph R, Ganesan NM (1998) Genetic variability and diversity for protein and calcium contents in finger millet Eleusine coracana L. Gaertn in relation to grain color. Plant Foods Hum Nutr 52:353–364
Pamidimarri DVNS, Chattopadhyay B, Reddy MP (2009) Genetic divergence and phylogenetic analysis of genus Jatropha based on nuclear ribosomal DNA ITS sequence. Mol Biol Rep 36:1929–1935
Kumar J, Verma V, Qazi GN, Gupta PK (2007) Genetic diversity in cymbopogon species using PCR-based functional markers. J Plant Biochem Biotechnol 16:119–122
Muthusamy S, Kanagarajan S, Ponnusamy S (2008) Efficiency of RAPD and ISSR markers system in accessing genetic variation of rice bean Vigna umbellata landraces. Electron J Biotechnol 11(3):1–10
Das S, Mishra RC, Rout GR, Aparajita S (2007) Genetic variability and relationships among thirty genotypes of finger millet Eleusine coracana L. Gaertn. using RAPD markers. J Biosci 621–2:116–122
Teulat B, Aldam C, Trehin R, Lepbrun P, Barker JHA, Arnold GM, Karp A, Baudouin L, Rognan F (2000) An analysis of genetic diversity in coconut Cocos nucifera populations from across the geographic range using sequence tagged microsatellites SSRs and AFLPs. Theor Appl Genet 100:764–771
Acknowledgments
The authors wish to acknowledge the Department of Biotechnology, Govt. of India for providing financial support in the form of Programme Support for research and development in Agricultural Biotechnology at G.B. Pant University of Agriculture and Technology, Pantnagar (Grant No. BT/PR7849/AGR/02/374/2006). The first author was supported by fellowship from DBT, India during the project programme. The authors thank the All India Co-ordinated Small Millets Improvement Project, ICAR, UAS, GKVK, Bangalore for providing the seed samples of germplasm analysed in present study. We are highly thankful to Dr. T. Mohapatra for his kind academic inputs in data analysis and interpretation of data.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Panwar, P., Saini, R.K., Sharma, N. et al. Efficiency of RAPD, SSR and Cytochrome P450 gene based markers in accessing genetic variability amongst finger millet (Eleusine coracana) accessions. Mol Biol Rep 37, 4075–4082 (2010). https://doi.org/10.1007/s11033-010-0067-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0067-5