Abstract
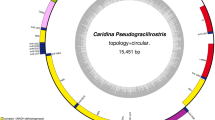
Given the commercial and ecological importance of the Asian paddle crab, Charybdis japonica, there is a clearly need for genetic and molecular research on this species. Here, we present the complete mitochondrial genome sequence of C. japonica, determined by the long-polymerase chain reaction and primer walking sequencing method. The entire genome is 15,738 bp in length, encoding a standard set of 13 protein-coding genes, two ribosomal RNA genes, and 22 transfer RNA genes, plus the putative control region, which is typical for metazoans. The total A+T content of the genome is 69.2%, lower than the other brachyuran crabs except for Callinectes sapidus. The gene order is identical to the published marine brachyurans and differs from the ancestral pancrustacean order by only the position of the tRNA His gene. Phylogenetic analyses using the concatenated nucleotide and amino acid sequences of 13 protein-coding genes strongly support the monophyly of Dendrobranchiata and Pleocyemata, which is consistent with the previous taxonomic classification. However, the systematic status of Charybdis within subfamily Thalamitinae of family Portunidae is not supported. C. japonica, as the first species of Charybdis with complete mitochondrial genome available, will provide important information on both genomics and molecular ecology of the group.





Similar content being viewed by others
References
Boore JL (1999) Animal mitochondrial genomes. Nucleic Acids Res 27:1767–1780. doi:10.1093/nar/27.8.1767
Parsons TJ, Coble MD (2001) Increasing the forensic discrimination of mitochondrial DNA testing through the analysis of the entire mitochondrial DNA genome. Croat Med J 42:304–309
Boore JL, Medina M, Rosenberg LA (2004) Complete sequences of two highly rearranged molluscan mitochondrial genomes, those of the scaphopod Graptacme eborea and of the bivalve Mytilus edulis. Mol Biol Evol 21:1492–1503. doi:10.1093/molbev/msh090
Macey JR, Papenfuss TJ, Kuehl JV, Fourcade HM, Boore JL (2004) Phylogenetic relationships among amphisbaenian reptiles based on complete mitochondrial genome sequences. Mol Phylogenet Evol 33:22–31. doi:10.1016/j.ympev.2004.05.003
Hua J, Li M, Dong P, Xie Q, Bu W (2009) The mitochondrial genome of Protohermes concolorus Yang et Yang 1988 (Insecta: Megaloptera: Corydalidae). Mol Biol Rep. doi:10.1007/s11033-008-9379-0 (in press)
Kim SR, Kim MI, Hong MY, Kim KY, Kang PD, Hwang JS, Han YS, Jin BR, Kim I (2009) The complete mitogenome sequence of the Japanese oak silkmoth, Antheraea yamamai (Lepidoptera: Saturniidae). Mol Biol Rep (in press). doi:10.1007/s11033-008-9393-2
Zhou Z, Huang Y, Shin F, Ye H (2009) The complete mitochondrial genome of Deracantha onos (Orthoptera: Bradyporidae). Mol Biol Rep 36:7–12. doi:10.1007/s11033-007-9145-8
Boore JL, Brown WM (1998) Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Curr Opin Genet Dev 8:668–674. doi:10.1016/S0959-437X(98)80035-X
Boore JL, Lavrov DV, Brown WM (1998) Gene translocation links insects and crustaceans. Nature 392:667–668. doi:10.1038/33577
Boore JL, Macey JR, Medina M (2005) Sequencing and comparing whole mitochondrial genomes of animals. Methods Enzymol 395:311–348. doi:10.1016/S0076-6879(05)95019-2
Place AR, Feng X, Steven CR, Fourcade HM, Boore JL (2005) Genetic markers in blue crabs (Callinectes sapidus). II. Complete mitochondrial genome sequence and characterization of genetic variation. J Exp Mar Biol Ecol 319:15–27. doi:10.1016/j.jembe.2004.03.024
Yamauchi MM, Miya MU, Nishida M (2003) Complete mitochondrial DNA sequence of the swimming crab, Portunus trituberculatus (Crustacea: Decapoda: Brachyura). Gene 311:129–135. doi:10.1016/S0378-1119(03)00582-1
Miller AD, Murphy NP, Burridge CP, Austin CM (2005) Complete mitochondrial DNA sequences of the decapod crustaceans Pseudocarcinus gigas (Menippidae) and Macrobrachium rosenbergii (Palaemonidae). Mar Biotechnol 7:339–349. doi:10.1007/s10126-004-4077-8
Sun H, Zhou K, Song D (2005) Mitochondrial genome of the Chinese mitten crab Eriocheir japonica sinenesis (Brachyura: Thoracotremata: Grapsoidea) reveals a novel gene order and two target regions of gene rearrangements. Gene 349:207–217. doi:10.1016/j.gene.2004.12.036
Segawa RD, Aotsuka T (2005) The mitochondrial genome of the Japanese freshwater crab, Geothelphusa dehaani (Crustacea: Brachyura): evidence for its evolution via gene duplication. Gene 355:28–39. doi:10.1016/j.gene.2005.05.020
Ng PKL, Guinot D, Davie PJF (2008) Systema Brachyuorum: part I. An annotated checklist of extant Brachyuran crabs of the world. Raffles Bull Zool 17:1–286
Wee DP, Ng PKL (1995) Swimming crabs of the genera Charybdis De Haan, 1833 and Thalamita Latreille, 1829 (Crustacea: Decapoda: Brachyura: Portunidae) from peninsular Malaysia and Singapore. Raffles Bull Zool (Suppl No) 1:1–128
Smith PJ, Webber WR, McVeagh SM, Inglis GJ, Gust N (2003) DNA, morphological identification of an invasive swimming crab Charybdis japonica (A. Milne-Edwards 1861) in New Zealand waters. N Zeal J Mar Fresh 37:753–762
Gust N, Inglis GJ (2006) Adaptive multi-scale sampling to determine an invasive crab’s habitat usage and range in New Zealand. Biol Invasions 8:339–353. doi:10.1007/s10530-004-8243-y
Chu KH, Tong J, Chan T-Y (1999) Mitochondrial cytochrome oxidase I sequence divergence in some Chinese species of Charybdis (Crustacea: Decapoda: Portunidae). Biochem Syst Ecol 27:461–468. doi:10.1016/S0305-1978(98)00115-X
Gao T, Zhang X, Watanabe S, Fu SR (2004) A preliminary study of mitochondrial DNA 12S rRNA gene sequence of Charybdis japonica and Thalamita prymna. J Ocean Univ China 34:43–47
Zhang S, Li XL, Cui ZX, Wang HX, Wang CL, Liu XL (2008) The Applications of mitochondrial DNA in phylogeny reconstruction and species identification of portunid crab. Mark Sci 32:9–18
Martin JW, Davis GE (2001) An updated classification of the recent crustacea. Natural History Museum of Los Angeles County, Los Angeles
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299
Lavrov DV, Brown WM, Boore JL (2004) Phylogenetic position of the Pentastomida and (pan)crustacean relationships. Proc R Soc Lond B Biol Sci 271:537–544. doi:10.1098/rspb.2003.2631
Boore JL, Brown WM (2000) Mitochondrial genomes of Galathealinum, Helobdella, and Platynereis: sequence and gene arrangement comparisons indicate that Pogonophora is not a phylum and Annelida and Arthropoda are not sister taxa. Mol Biol Evol 17:87–106
Cheng S, Chang SY, Gravitt P, Respess R (1994) Long PCR. Nature 369:684–685. doi:10.1038/369684a0
Ewing B, Green P (1998) Basecalling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194. doi:10.1101/gr.8.3.186
Ewing B, Hillier L, Wendl M, Green P (1998) Basecalling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8:175–185. doi:10.1101/gr.8.3.175
Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202. doi:10.1101/gr.8.3.195
Wyman SK, Jansen RK, Boore JL (2004) Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255. doi:10.1093/bioinformatics/bth352
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi:10.1093/nar/25.5.955
Lohse M, Drechsel O, Bock R (2007) OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet 52:267–274. doi:10.1007/s00294-007-0161-y
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599. doi:10.1093/molbev/msm092
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. doi:10.1093/nar/25.24.4876
Xia X, Xie Z, Salemi M, Chen L, Wang Y (2003) An index of substitution saturation and its application. Mol Phylogenet Evol 26:1–7. doi:10.1016/S1055-7903(02)00326-3
Xia X, Xie Z (2001) DAMBE: software package for data analysis in molecular biology and evolution. J Hered 92:371–373. doi:10.1093/jhered/92.4.371
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi:10.1080/10635150390235520
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. doi:10.1093/bioinformatics/17.8.754
Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818. doi:10.1093/bioinformatics/14.9.817
Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105. doi:10.1093/bioinformatics/bti263
Shen X, Ren J, Cui Z, Sha Z, Wang B, Xiang J, Liu B (2007) The complete mitochondrial genomes of two common shrimps (Litopenaeus vannamei and Fenneropenaeus chinensis) and their phylogenomic considerations. Gene 403:98–109. doi:10.1016/j.gene.2007.06.021
Shen X, Sun M, Wu Z, Tian M, Cheng H, Zhao F, Meng X (2009) The complete mitochondrial genome of the ridgetail white prawn Exopalaemon carinicauda Holthuis, 1950 (Crustacean: Decapoda: Palaemonidae) revealed a novel rearrangement of tRNA gene. Gene 437:1–8. doi:10.1016/j.gene.2009.02.014
Wolstenholme DR (1992) Animal mitochondrial DNA: structure and evolution. Int Rev Cytol 141:173–216. doi:10.1016/S0074-7696(08)62066-5
Ojala D, Montoya J, Attardi G (1981) tRNA punctuation model of RNA processing in human mitochondria. Nature 290:470–474. doi:10.1038/290470a0
Yang JS, Nagasawa H, Fujiwara Y, Tsuchida S, Yang WJ (2008) The complete mitochondrial genome sequence of the hydrothermal vent galatheid crab Shinkaia crosnieri (Crustacea: Decapoda: Anomura): a novel arrangement and incomplete tRNA suite. BMC Genomics 9:257. doi:10.1186/1471-2164-9-257
Hickerson MJ, Cunningham CW (2000) Dramatic mitochondrial gene rearrangements in the hermit crab Pagurus longicarpus (Crustacea, anomura). Mol Biol Evol 17:639–644
Miller AD, Nguyen TT, Burridge CP, Austin CM (2004) Complete mitochondrial DNA sequence of the Australian freshwater crayfish, Cherax destructor (Crustacea: Decapoda: Parastacidae): a novel gene order revealed. Gene 331:65–72. doi:10.1016/j.gene.2004.01.022
Boas JEV (1880) Studier over Decapodernes Slaegtskabsforhold. Kongel Danske Videnske Selsk Skr 6:25–210
Felgenhauer BE, Abele LG (1983) Phylogenetic relationships among shrimp-like decapods. Crustac Issues 1:291–311
Abele LG, Felgenhauer BE (1986) Phylogenetic and phenetic relationships among the lower Decapoda. J Crustac Biol 6:385–400. doi:10.2307/1548179
Abele LG (1991) Comparison of morphological and molecular phylogeny of the Decapoda. Mem Queensl Mus 31:101–108
Burkenroad MD (1963) The evolution of the Eucarida (Crustacea, Eumalacostraca), in relation to the fossil record. Tulane Stud Geol 2:1–17
Burkenroad MD (1981) The higher taxonomy and evolution of Decapoda (Crustacea). Trans San Diego Soc Nat Hist 19:251–268
Tsang LM, Ma KY, Ahyong ST, Chan T-Y, Chu KH (2008) Phylogeny of Decapoda using two nuclear protein-coding genes: origin and evolution of the Reptantia. Mol Phylogenet Evol 48:359–368. doi:10.1016/j.ympev.2008.04.009
Scholtz G, Richter S (1995) Phylogenetic systematics of the reptantian Decapoda (Crustacea, Malacostraca). Zool J Linn Soc 113:289–328. doi:10.1111/j.1096-3642.1995.tb00936.x
Schram FR (2001) Phylogeny of decapods: moving towards a consensus. Hydrobiologia 449:1–20. doi:10.1023/A:1017543712119
Dixon CJ, Ahyong ST, Schram FR (2003) A new hypothesis of decapod phylogeny. Crustaceana 76:935–975. doi:10.1163/156854003771997846
Miller AD, Austin CM (2006) The complete mitochondrial genome of the mantid shrimp Harpiosquilla harpax, and a phylogenetic investigation of the Decapoda using mitochondrial sequences. Mol Phylogenet Evol 38:565–574. doi:10.1016/j.ympev.2005.10.001
Ivey JL, Santos SR (2007) The complete mitochondrial genome of the Hawaiian anchialine shrimp Halocaridina rubra Holthuis, 1963 (Crustacea: Decapoda: Atyidae). Gene 394:35–44. doi:10.1016/j.gene.2007.01.009
Stephenson W, Campbell B (1960) The Australian portunids (Crustacea: Portunidae) IV: remaining genera. Aust J Mar Freshwater Res 11:73–122. doi:10.1071/MF9600073
Dai AY, Yang SL, Song YZ, Chen GX (1986) Crabs of the China seas. China Ocean Press, Beijing
Apel M, Spiridonov VA (1998) Taxonomy and zoogeography of the portunid crabs (Crustacea: Decapoda: Brachyura: Portunidae) of the Arabian Gulf and adjacent waters. Fauna Arabia 17:159–331
Ng PKL, Wang C-H, Ho P-H, Shih H-T (2001) An annotated checklist of brachyuran crabs from Taiwan (Crustacea: Decapoda). Natl Taiwan Mus Spec Publ Ser 11:1–86
Acknowledgments
We thank Prof. Xinzheng LI for specimen identification and Mr. Ping ZHANG for assistance in bioinformatics analyses. This research was supported by the National Natural Science Foundation of China (no. 40676085) and Chinese National ‘863’ Project (no. 2006AA10A406).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, Y., Cui, Z. Complete mitochondrial genome of the Asian paddle crab Charybdis japonica (Crustacea: Decapoda: Portunidae): gene rearrangement of the marine brachyurans and phylogenetic considerations of the decapods. Mol Biol Rep 37, 2559–2569 (2010). https://doi.org/10.1007/s11033-009-9773-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-009-9773-2