Abstract
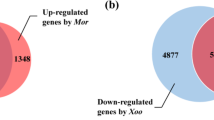
In order to understand the mechanism of the strong resistance of Oryza granulata to Xanthomonas oryzae pv oryzae (Xoo), cDNA microarrays containing 2,436 cDNA clones of Oryza granulata derived from Suppression subtractive library and cDNA library were constructed and genome expression patterns after inoculating Xoo were investigated. Three hundred and 83 clones were up-regulated, 836 clones were down-regulated after pathogen infection. Approximately 800 clones were sequenced and BLAST search were carried out. There are no homologous sequences for 35 clones of them. The functions of the homologous sequences for most clones are unknown. The known functions of the homologous sequences involved in photosynthesis, respiration, material transport, signal transduction, pathogenesis-related proteins, transcription factors, the active oxygen scavenging system and so on. The putative functions of them in responding to Xoo were discussed.










Similar content being viewed by others
References
Peng SQ, Wei ZS, Mao CX (1981) Identification of wild rice resistance: O. granulata with a highly resistant to Xoo. Hunan Agr Sci 5:47–48
Zhang Q, Wang CL, Shi AN et al (1994) Evaluation of resistance to bacterial blight (Xanthomonas oryzae pv. oryzae) in wild rice species. Sci Agr Sin 27:1–9
Cheng ZQ, Yan HJ, Geng XS, Yin FY, Sun YD, Zhang C, Huang XQ (2008) Identification of Oryza granulata for resistance to Xanthomonas oryzae pv. oryzae and observation of leaf tissue. Acta Phytopathol Sin 38:582–591
HuangPu WG, Ying CB, Qiu RJ et al (2001) Studies on 4th generation character of wild rice and cultivated rice somatic hybridization. J Henan Vocat Coll 29:12–14
Zhu YS, Chen BT, Yu SW (2004) Transferring the resistance of O. granulata to Xoo by asymmetric somatic hybrids. Chin Sci Bull 49:1395–1398
Song WY, Wang GL, Chen LL, Kim HS et al (1995) A receptor kinase-like protein encoded by the rice disease resistance gene Xa21. Science 270:1772–1804
Yoshimura S, Yamanouchi U, Kayayose Y et al (1998) Expression of Xa1, a bacterial blight-resistance gene in rice is induced by bacterial inoculation. Proc Natl Acad Sci USA 95:1663–1668
Sun X, Cao Y, Yang Z, Xu C, Li X, Wang S, Zhang Q (2004) Xa26 a gene conferring resistance to Xanthomouas oryzae Pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J 37:517–527
Iyer AS, Mccouch SR (2004) The rice bacterial blight resistance gene Xa5 encodes a novel form of disease resistance. Mol Plant Microbe Interact 17:1348–1354
Gu K, Tian D, Yang F et al (2004) High-resolution genetic mapping of Xa27(t) a new bacterial blight resistance gene in rice Oryza Sativa L. Theor Appl Genet 108:800–807
Chu ZH, Fu BY, Yang H et al (2006) Targeting xa13, a recessive gene for bacterial blight resistance in rice. Theor Appl Genet 112:455–461
Qian J, Cheng ZQ, Yang MZ, Liu JM, Wu CJ, Huang XQ (2005) Isolation and sequence analysis of the Xa21 gene exon II homologs from different species of wild rice in Yunnan. Hereditas 27:382–386
Xiong ZY, Tan GX, You AQ (2004) Comparative physical mapping of rice BAC clones linked to resistance genes Glh, Bph-3 and xa-5 in Oryza sativa and O. granulate. Chin Sci Bull 49:252–257
Kottapalli KR, Rakwal R, Satoh K et al (2007) Transcriptional profiling of indica rice cultivar IET8585 (Ajaya) infected with bacterial leaf blight pathogen Xanthomonas oryzae pv oryzae. Plant Physiol Biochem 45:834–850
Lia Q, Chena F, Sun L et al (2006) Expression profiling of rice genes in early defense responses to blast and bacterial blight pathogens using cDNA microarray. Physiol Mol Plant Pathol 68:51–60
Zhou B, Peng KM, Chu ZH (2002) The defense-responsive genes showing enhanced and repressed expression after pathogen infection in rice (Oryza sativa L.). Sci Chin (Series C) 45(44):9–467
Qian J, Zhang S, Ji G (2003) Evaluation of resistance to bacterial blight (Xanthomonas oryzae pv. oryzae) in Yunnan rice cultivars. J Yunnan Agric Univ 18:239–245
Chu ZH, Peng KM, Zhang LD et al (2002) Construction and characterization of a normalized cDNA library of rice all life. Chin Sci Bull 47:1656–1662
Lan WZ, He GC, Wu SJ, Qin R (2006) Comparative analysis of Oryza sativa, O. officinalis and O. meyeriana genome with C0t–1 DNA and genomic DNA. Chin J Agric Sci 39:1083–1090
Wang HH, Cao CS, Kang J, Zeng FH (2002) Systemic resistance of rice to bacterial blight induced by salicylic acid and changes in activities of some enzymes in untreated leaves. Chin J Rice Sci 16:252–256
Gagne JM, Smalle J, Gingerich DJ, Walker JM, Yoo S-D, Yanagisawa S, Vierstra RD (2004) Arabidopsis EIN3-binding F-box and form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc Natl Acad Sci USA 101:6803–6808
Gagne JM, Downes BP, Shiu S-H, Durski AM, Vierstra RD (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99:11519–11524
Gray WM, Kepinski S, Rouse D et al (2001) Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414:271–276
Gray WM et al (2002) Role of the Arabidopsis RING2H2 protein RBX1 in RUB modification and SCF function. Plant Cell 14:2137–2144
Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435:441–445
Kitagawa K, Hieter P (2001) Evolutionary conservation between budding yeast and human kinetochores. Nat Rev Mol Cell Biol 2:678–687
Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JDG, Parker JE (2002) Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295:2077–2080
Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P (2002) The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295:2073–2076
Liu Y, Schiff M, Serino G, Deng X-W, Dinesh-Kumar SP (2002) Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to tobacco mosaic virus. Plant Cell 14:1483–1496
Tor M, Gordon P, Cuzick A, Eulgem T, Sinapidou E, Mert F, Can C, Dangl JL, Holub EB (2002) Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew (Peronospora parasitica) resistance genes. Plant Cell 14:993–1003
Schwechheimer C, Deng X-W (2001) COP9 signalosome revisited: a novel mediator of protein degradation. Trends Cell Biol 11:420–426
Kim HS, Delaney TP (2002) Arabidopsis SON1 is an F-box protein that regulates a novel induced defense response independent of both salicylic acid and systemic acquired resistance. Plant Cell 14:1469–1482
Boyes DC, Nam J, Dangl JL (1998) The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc Natl Acad Sci USA 95:15849–15854
Schwechheimer C, Serino G, Deng X-W (2002) Multiple ubiquitin ligase-mediated processes require COP9 signalosome and AXR1 function. Plant Cell 14:2553–2563
Gonzalez-Lamothe R, Tsitsigiannis DI, Ludwig AA, Panicot M, Shirasu K, Jones JDG (2006) The U-Box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resis-tance genes in tobacco and tomato. Plant Cell 18:1067–1083
Yang CW, Gonzalez-Lamothe R, Ewan RA, Rowland O, Yoshioka H, Shenton M, Ye H (2006) A. Sadanan-dom, The E3 ubiquitin ligase activity of Arabidopsis PLANTU-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18:1084–1098
Shirasu K, Schulze-Lefert P (2003) Complex formation, promiscuity and multi-functionality: protein interactions in disease-resistance pathways. Trends Plant Sci 8:252–258
Dubacq C, Guerois R, Courbeyrette R, Kitagawa K, Mann C (2002) Sgt1p contributes to cyclic AMP pathway activity and physically interacts with the adenylyl cyclase Cyr1p/Cdc35p in budding yeast. Eukaryot Cell 1:568–582
Garcia-Hernandez M, Murphy A, Taiz L (1998) Metallothioneins 1 and 2 have distinct but overlapping expression patterns in Arabidopsis. Plant Physiol 118:387–397
Molina A, Garcia OF (1997) Enhanced tolerance to bacterial pathogens caused by the transgenic expression of barley lipid transfer protein LTP2. Plant J 12:669–675
Chassot C, Nawrath C, Metraux JP (2007) Cuticular defects lead to full immunity to a major plant pathogen. Plant J 49:972–980
Maldonado AM, Doerner P, Dixon RA, Lamb CJ, Cameron RK (2002) A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419:399–403
Buhot N, Douliez JP, Jacquemard A et al (2001) A lipid transfer protein binds to a receptor involved in the control of plant defenses. FEBS Lett 509:27–30
Sonnewald U, Sturm A, Chrispeels MJ, Willmitzer L (1989) Targeting and glycosylation of patatin, the major potato tuber protein in leaves of transgenic tobacco. Planta 179:171–180
Twell D, Ooms G (1988) Structural diversity of the patatin gene family in potato cv. Desiree. Mol Gen Genet 212:325–336
Koda Y, Kikuta Y (1994) Wound-induced accumulation of jasmolic acid in tissues of potato tubers. Plant Cell Physiol 35:751–756
Sembder G, Parthier B (1993) The biochemistry and the physiological and molecular action of jasmonates. Plant Mol Biol 44:569–589
Si HJ, Liu J, Xie CH (2004) Transformation of potato using an antisense class I patatin genes and its effect on microtuber formation. J Agric Biotechnol 12:147–151
Romeis T, Ludwig AA, Martin R, Jones JD (2001) Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J 20:5556–5567
Romeis T, Piedras P, Jones JD (2000) Resistance gene-dependent activation of a calcium-dependent protein kinase in the plant defense response. Plant Cell 12:803–816
Romeis T, Piedras P, Zhang S, Klessig DF, Hirt H, Jones JD (1999) Rapid Avr9-and Cf-9-dependent activation of MAP kinases in tobacco cell cultures and leaves: convergence of resistance gene, elicitor, wound, and salicylate responses. Plant Cell 11:273–287
Zhang S, Klessig DF (1998) Resistance gene N-mediated de novo synthesis and activation of a tobacco mitogen-activated protein kinase by tobacco mosaic virus infection. Proc Natl Acad Sci USA 95:7433–7438
Zhang S, Liu Y (2001) Activation of salicylic acid-induced protein kinase, a mitogen-activated protein kinase, induces multiple defense responses in tobacco. Plant Cell 13:1877–1889
Yang KY, Liu Y, Zhang S (2001) Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc Natl Acad Sci USA 98:741–746
Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE, Sharma SB, Klessig DF, Martienssen R, Mattsson O, Jensen AB, Mundy J (2000) Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103:1111–1120
Wiermer M, Feys BJ, Parker JE (2005) Plant immunity: the EDS1 regulatory node. Curr Opin Plant Biol 8:383–389
Thomma BPHJ, Penninckx IAMA, Broekaert WF, Cammue BPA (2001) The complexity of disease signaling in Arabidopsis. Curr Opin Immunol 13:63–68
Thomma BPHJ, Nelissen L, Eggermont K et al (1999) Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to fungal Alternaria brassicicola. Plant J 19:163–171
Glazebrook J, Zook M, Mert F et al (1997) Phytoalexin deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics 146:381–392
Kim YC, Kim SY, Paek KH et al (2006) Suppression of CaCYP1, a novel cytochrome P450 gene, compromises the basal pathogen defense response of pepper plants. Biochem Biophys Res 345:638–645
Schenk H, Klein M, Erdbrügger W et al (1994) Distinct effects of thioredoxin and antioxidants on the activation of transcription factors NF-kappa B and AP-1. Proc Natl Acad Sci USA 91:1672–1676
Saitoh M, Nishitoh H, Fujii M et al (1998) Mammalian thioredox in is a direct inhibitor of apoptosis signal regulating kinase (ASK) l. EMBO J 17:2596–2606
Cosgrove DJ (2000) New genes and new biological roles for expansins. Curr Opin Plant Biol 3:73–78
Bouzareloua D, Billinia M, Roumeliotia K, Sophianopoulou V (2008) EglD, a putative endoglucanase, with an expansin like domain is localized in the conidial cell wall of Aspergillus nidulans. Fungal Genet Biol 45:839–850
Liu QP, Feng Y, Dong H (2005) Insilico cloning of mSHMT gene from rice (Oryza sativa L.). Chin J Bioinformatics 3:5–9
Acknowledgments
This research was funded by the National Science Foundation of China (No. 30460019 and 30660090), and Key Project of Natural Science Foundation of Yunnan Province (No. 2008C004).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Luo, Y., Lv, GL., Wu, WT. et al. Analysis of genome expression in the response of Oryza granulata to Xanthomonas oryzae pv oryzae . Mol Biol Rep 37, 875 (2010). https://doi.org/10.1007/s11033-009-9694-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-009-9694-0