Abstract
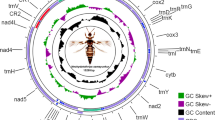
The first complete mitochondrial genome of dobsonfly Protohermes concolorus Yang et Yang, 1988 (Megaloptera: Corydalidae) was sequenced in this study. The genome was a circular molecule of 15,851 bp containing the typical 37 genes that arranged in the same order as that of the putative ancestor of hexapods. Sequences overlaps were observed between several neighbor genes, which made the genome relatively compact. The tRNA-Ser (GCT) could not be folded into typical secondary structure because its DHU arm was replaced with a simple loop. Six of the 13 protein genes were terminated with a single T adjacent to a downstream tRNA gene in the same strand. The variation of GC content caused the different nucleotide substitution patterns of the protein genes. The genome was AT-biased with a total A + T content of 75.83% which was also demonstrated by the codon usage. The control region was the most AT-rich region with a sub-region of even higher A + T content. Protein genes of two strands presented opposite CG-skew trends which was also reflected by the codon usage. For most of the amino acids, the protein coding sequences did not prefer to use the cognate codons of corresponding tRNAs and the codon usage of the protein genes was not random. The variation of nucleotide substitution patterns of protein genes was significantly correlated with the GC content. The phylogenetic analyses based on all the 13 protein genes showed that Megaloptera was the sister group of other holometabolous insects except Coleoptera.




Similar content being viewed by others
References
Avise JA, Aronold J, Ball RM, Bermingham E, Neigel JE, Reeb CA et al (1987) Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu Rev Ecol Syst 18:489–522
Simon C, Frati F, Beckenbach A, Cresp B, Liu H, Flook P (1994) Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am 87:651–701
Boore JL, Brown WM (1998) Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Curr Opin Genet Dev 8(6):668–674. doi:10.1016/S0959-437X(98)80035-X
Larget B, Simon DL, Kadane JB (2002) Bayesian phylogenetic inference from animal mitochondrial genome arrangements. J R Stat Soc Ser B Stat Methodol 64:681–693. doi:10.1111/1467-9868.00356
Grimaldi D, Engel MS (2005) Evolution of the insects. Cambridge University Press, New York, NY
Cameron SL, Beckenbach AT, Dowton M, Whiting MF (2006) Evidence from mitochondrial genomics on interordinal relationships in insects. Arthropod Syst Phylogeny 64(1):27–34
Wheeler W (2001) Homology and the optimization of DNA sequence data. Cladistics 17(1):S3–S11. doi:10.1111/j.1096-0031.2001.tb00100.x
Whiting MF (2002) Phylogeny of the holometabolous insect orders based on 18S ribosomal DNA: when bad things happen to good data. In: DeSalle R, Giribet G, Wheeler W (eds) EXS (Basel) molecular systematics and evolution: theory and practice. Birkhaeuser Boston Birkhaeuser Publishing Ltd, Cambridge, pp 69–83
Whiting MF, Carpenter JC, Wheeler QD, Wheeler WC (1997) The Strepsiptera problem: phylogeny of the holometabolous insect orders inferred from 18S and 28S ribosomal DNA sequences and morphology. Syst Biol 46(1):1–68. doi:10.2307/2413635
Reineke A, Karlovsky P, Zebitz CP (1998) Preparation and purification of DNA from insects for AFLP analysis. Insect Mol Biol 7(1):95–99. doi:10.1046/j.1365-2583.1998.71048.x
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Acids Symp Ser 41:95–98
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25(24):4876–4882. doi:10.1093/nar/25.24.4876
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25(5):955–964. doi:10.1093/nar/25.5.955
Peden JF (1999) Analysis of codon usage. University of Nottingham, UK
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17(8):754–755. doi:10.1093/bioinformatics/17.8.754
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14:814–818. doi:10.1093/bioinformatics/14.9.817
Swofford DL (1998) PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Assocates Inc, Sunderland, MA
Guindon S, Lethiec F, Duroux P, Gascuel O (2005) PHYML online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res 33(Web Server issue):W557–W559
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi:10.2307/2408678
Lavrov DV, Brown WM, Boore JL (2000) A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. Proc Natl Acad Sci USA 97(25):13738–13742. doi:10.1073/pnas.250402997
Stewart JB, Beckenbach AT (2005) Insect mitochondrial genomics: the complete mitochondrial genome sequence of the meadow spittlebug Philaenus spumarius (Hemiptera: Auchenorrhyncha: Cercopoidae). Genome 48(1):46–54. doi:10.1139/g04-090
Adams KL, Palmer JD (2003) Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol 29(3):380–395. doi:10.1016/S1055-7903(03)00194-5
Podsiadlowski L, Braband A, Mayer G (2008) The complete mitochondrial genome of the onychophoran Epiperipatus biolleyi reveals a unique transfer RNA set and provides further support for the ecdysozoa hypothesis. Mol Biol Evol 25(1):42–51. doi:10.1093/molbev/msm223
Boore JL (2001) Complete mitochondrial genome sequence of the polychaete annelid Platynereis dumerilii. Mol Biol Evol 18(7):1413–1416
Boore JL (2006) The complete sequence of the mitochondrial genome of Nautilus macromphalus (Mollusca: Cephalopoda). BMC Genom 7:182. doi:10.1186/1471-2164-7-182
Boore JL, Brown WM (2000) Mitochondrial genomes of Galathealinum, Helobdella, and Platynereis: sequence and gene arrangement comparisons indicate that Pogonophora is not a phylum and Annelida and Arthropoda are not sister taxa. Mol Biol Evol 17(1):87–106
Shao R, Campbell NJ, Schmidt ER, Barker SC (2001) Increased rate of gene rearrangement in the mitochondrial genomes of three orders of hemipteroid insects. Mol Biol Evol 18(9):1828–1832
Wang X, Lavrov DV (2007) Mitochondrial genome of the homoscleromorph Oscarella carmela (Porifera, Demospongiae) reveals unexpected complexity in the common ancestor of sponges and other animals. Mol Biol Evol 24(2):363–373. doi:10.1093/molbev/msl167
Ojala D, Montoya J, Attardi G (1981) tRNA punctuation model of RNA processing in human mitochondria. Nature 290(5806):470–474. doi:10.1038/290470a0
Stanton DJ, Daehler LL, Moritz CC, Brown WM (1994) Sequences with the potential to form stem-and-loop structures are associated with coding-region duplications in animal mitochondrial DNA. Genetics 137(1):233–241
Rand D (2001) Mitochondrial genomics flies high. Trends Ecol Evol 16(1):2–4. doi:10.1016/S0169-5347(00)02036-X
Rand DM (1993) Endotherms, ectotherms, and mitochondrial genome-size variation. J Mol Evol 37(3):281–295. doi:10.1007/BF00175505
Cook CE (2005) The complete mitochondrial genome of the stomatopod crustacean Squilla mantis. BMC Genom 6:105. doi:10.1186/1471-2164-6-105
Wolstenholme DR (1992) Animal mitochondrial DNA: structure and evolution. Int Rev Cytol 141:173–216. doi:10.1016/S0074-7696(08)62066-5
Thao ML, Baumann L, Baumann P (2004) Organization of the mitochondrial genomes of whiteflies, aphids, and psyllids (Hemiptera, Sternorrhyncha). BMC Evol Biol 4:25. doi:10.1186/1471-2148-4-25
Kolpakov R, Bana G, Kucherov G (2003) mreps: Efficient and flexible detection of tandem repeats in DNA. Nucleic Acids Res 31(13):3672–3678. doi:10.1093/nar/gkg617
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402. doi:10.1093/nar/25.17.3389
Perna NT, Kocher TD (1995) Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol 41(3):353–358. doi:10.1007/BF01215182
Hassanin A, Leger N, Deutsch J (2005) Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of metazoa, and consequences for phylogenetic inferences. Syst Biol 54(2):277–298. doi:10.1080/10635150590947843
Gibson A, Gowri-Shankar V, Higgs PG, Rattray M (2005) A comprehensive analysis of mammalian mitochondrial genome base composition and improved phylogenetic methods. Mol Biol Evol 22(2):251–264. doi:10.1093/molbev/msi012
Xia X (1996) Maximizing transcription efficiency causes codon usage bias. Genetics 144(3):1309–1320
Marashi SA, Najafabadi HS (2004) How reliable re-adjustment is: correspondence regarding A. Fuglsang, The ‘effective number of codons’ revisited. Biochem Biophys Res Commun 324(1):1–2. doi:10.1016/j.bbrc.2004.08.213
Wright F (1990) The `effective number of codons’ used in a gene. Gene 87(1):23–29. doi:10.1016/0378-1119(90)90491-9
Kukalova-Peck J (1991) Fossil history and the evolution of hexapod structures. In: CSIRO (ed) The insects of Australia, vol 1, 2nd edn. Melbourne University Press, Carlton, pp 141–179
Boudreaux HB (1979) Arthropod phylogeny with special reference to insects. Wiley, New York
Kristensen NP (1991) Phylogeny of extant hexapods. In: CSIRO (ed) The insects of australia, vol 1, 2nd edn. Melbourne University Press, Carlton, pp 125–140
Acknowledgments
We thank Prof. Ding Yang and Dr. Xingyue Liu, China Agricultural University, for identifying the species of Megaloptera. This project was supported by Natural Science Foundation of China (No. 30725005 and No. J0630963), and by Ministry of Education of China (No. 20050055027).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hua, J., Li, M., Dong, P. et al. The mitochondrial genome of Protohermes concolorus Yang et Yang 1988 (Insecta: Megaloptera: Corydalidae). Mol Biol Rep 36, 1757–1765 (2009). https://doi.org/10.1007/s11033-008-9379-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-008-9379-0