Abstract
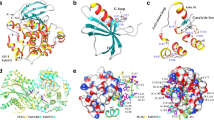
The tetratricopeptide repeat (TPR) proteins found in all kingdoms of life can mediate protein-protein interactions in a variety of biological systems through binding to specific peptide ligands, such as cell cycle control, transcription, protein transport and protein folding. Although the proteins are ubiquitous, no comprehensive overview of them in Arabidopsis thaliana (At), Oryza sativa (Os), Zea mays (Zm) and Populus trichocarpa (Pt) has been available in the literature till now. Through whole genome investigation, 177 Arabidopsis, 216 rice, 211 maize and 243 poplar TPR genes were identified and categorized into 28 subfamilies and 11 groups based on their domain compositions and phylogenetic relationships, respectively. Phylogenetic analysis revealed that most genes in the same subfamilies are classified into at least two groups, implying that TPR proteins have acquired functional diversity by extensive domain shuffling and/or emerged multiple times independently during evolution. Structural analysis of ZmTPR080 showed that eight TPR motifs as two anti-parallel α-helices form an amphipathic groove capable of accepting a target protein peptide. Similar expression patterns across development stages suggested functional conservation between homologous pairs, for example, AtTPR050/ZmTPR049 may have similar functions in the regulation of flowering. Under drought stress, 26 OsTPRs showed strong alterations of their expression levels in rice leaf, while 10 and 49 ZmTPRs were significantly differentially expressed in maize leaf and cob, respectively.




Similar content being viewed by others
References
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201
Aviezerhagai K, Skovorodnikova J, Galigniana MD et al (2007) Arabidopsis immunophilins ROF1 (AtFKBP62) and ROF2 (AtFKBP65) exhibit tissue specificity, are heat-stress induced, and bind HSP90. Plant Mol Biol 63:237–255
Bell SA, Hunt AG (2010) The Arabidopsis ortholog of the 77 kDa subunit of the cleavage stimulatory factor (AtCstF-77) involved in mRNA polyadenylation is an RNA-binding protein. FEBS Lett 584:1449–1454
Bhuiyan NH, Friso G, Poliakov A et al (2015) MET1 is a Thylakoid-associated TPR protein involved in Photosystem II Supercomplex formation and repair in Arabidopsis. Plant Cell Online 27:262–285
Champion EA, Lane BH, Jackrel ME et al (2008) A direct interaction between the Utp6 half-a-Tetratricopeptide repeat domain and a specific peptide in Utp21 is essential for efficient pre-rRNA processing. Mol Cell Biol 28:6547–6556
Chen L, Dodd IC, Davies WJ, Wilkinson S (2013) Ethylene limits abscisic acid- or soil drying-induced stomatal closure in aged wheat leaves. Plant Cell Environ 36:1850–1859
Chewawiwat N, Yano M, Terada K et al (1999) Characterization of the novel mitochondrial protein import component, Tom34, in mammalian cells. J Biochem 125:721–727
Christians MJ, Gingerich DJ, Hansen M et al (2009) The BTB ubiquitin ligases ETO1, EOL1 and EOL2 act collectively to regulate ethylene biosynthesis in Arabidopsis by controlling type-2 ACC synthase levels. Plant J 57:332–345
Dühring U, Irrgang KD, Lünser K et al (2006) Analysis of photosynthetic complexes from a cyanobacterial ycf37 mutant. Biochim Biophys Acta 1757:3–11
Dühring U, Ossenbühl F, Wilde A (2007) Late assembly steps and dynamics of the Cyanobacterial Photosystem I. J Biol Chem 282:10915–10921
El Zawily AM, Schwarzlander M, Finkemeier I et al (2014) FRIENDLY regulates mitochondrial distribution, fusion, and quality control in Arabidopsis. Plant Physiol 166:808–828
Felder S, Meierhoff K, Sane AP et al (2001) The nucleus-encoded HCF107 gene of Arabidopsis provides a link between Intercistronic RNA processing and the accumulation of translation-competent psbH transcripts in chloroplasts. Plant Cell 13:2127–2141
Feldheim D, Schekman R (1994) Sec72p contributes to the selective recognition of signal peptides by the secretory polypeptide translocation complex. J Cell Biol 126:935–943
Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:29–37
Gatto GJ, Geisbrecht BV, Gould SJ, Berg JM (2000) Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat Struct Biol 7:1091–1095
Golovkin M, Reddy ASN (2003) A calmodulin-binding protein from Arabidopsis has an essential role in pollen germination. Proc Natl Acad Sci U S A 100:10558–10563
Gray WM, Muskett PR et al (2003) Arabidopsis SGT1b is required for SCF(TIR1)-mediated auxin response. Plant Cell 15:1310–1319
Greenboimwainberg Y, Maymon I, Borochov R et al (2005) Cross talk between Gibberellin and Cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in Cytokinin Signaling. Plant Cell 17:92–102
Haucke V, Horst M, Schatz G, Lithgow T (1996) The Mas20p and Mas70p subunits of the protein import receptor of yeast mitochondria interact via the tetratricopeptide repeat motif in Mas20p: evidence for a single hetero-oligomeric receptor. EMBO J 15:1231–1237
He Y, Doyle MR, Amasino RM (2004) PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev 18:2774–2784
Heyman J, De Veylder L (2012) The anaphase-promoting complex/Cyclosome in control of plant development. Mol Plant 5:1182–1194
Johnson BD, Schumacher RJ, Ross ED, Toft DO (1998) Hop modulates Hsp70/Hsp90 interactions in protein folding. J Biol Chem 273:3679–3686
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066
Kauss D, Bischof S, Steiner S et al (2012) FLU, a negative feedback regulator of tetrapyrrole biosynthesis, is physically linked to the final steps of the mg++−branch of this pathway. FEBS Lett 586:211–216
Kumar A, Roach C, Hirsh IS et al (2001) An unexpected extended conformation for the third TPR motif of the peroxin PEX5 from Trypanosoma brucei. J Mol Biol 307:271–282
Kwon S, Kim SH, Bhattacharjee S et al (2009) SRFR1, a suppressor of effector-triggered immunity, encodes a conserved tetratricopeptide repeat protein with similarity to transcriptional repressors. Plant J 57:109–119
Lakhssassi N, Doblas VG, Rosado A et al (2012) The Arabidopsis TETRATRICOPEPTIDE THIOREDOXIN-LIKE gene family is required for osmotic stress tolerance and male Sporogenesis. Plant Physiol 158:1252–1266
Legrain P, Choulika A (1990) The molecular characterization of PRP6 and PRP9 yeast genes reveals a new cysteine/histidine motif common to several splicing factors. EMBO J 9:2775–2781
Li Z, Xu Z, He G et al (2012) A mutation in Arabidopsis BSK5 encoding a brassinosteroid-signaling kinase protein affects responses to salinity and abscisic acid. Biochem Biophys Res Commun 426:522–527
Li J, Liu J, Wang G et al (2015a) A chaperone function of NO CATALASE ACTIVITY1 is required to maintain catalase activity and for multiple stress responses in Arabidopsis. Plant Cell 27:908–925
Li N, Chen Y, Ding Z et al (2015b) Nonuniform gene expression pattern detected along the longitudinal axis in the matured rice leaf. Sci Rep 5:8015
Lin Z, Arcigareyes L, Zhong S et al (2008) SlTPR1, a tomato tetratricopeptide repeat protein, interacts with the ethylene receptors NR and LeETR1, modulating ethylene and auxin responses and development. J Exp Bot 59:4271–4287
Lin Z, Ho C, Grierson D (2009) AtTRP1 encodes a novel TPR protein that interacts with the ethylene receptor ERS1 and modulates development in Arabidopsis. J Exp Bot 60:3697–3714
Luo W, Ji Z, Pan Z et al (2013) The conserved Intronic cleavage and Polyadenylation site of CstF-77 gene imparts control of 3′ end processing activity through feedback autoregulation and by U1 snRNP. PLoS Genet 9:e1003613
Nasmyth K, Peters J, Uhlmann F (2000) Splitting the chromosome: cutting the ties that bind sister chromatids. Science 288:1379–1384
Naver H, Boudreau E, Rochaix J (2001) Functional studies of Ycf3 its role in assembly of Photosystem I and interactions with some of its subunits. Plant Cell 13:2731–2745
Park S, Khamai P, Garciacerdan JG, Melis A (2007) REP27, a tetratricopeptide repeat nuclear-encoded and chloroplast-localized protein, functions in D1/32-kD reaction center protein turnover and photosystem II repair from photodamage. Plant Physiol 143:1547–1560
Peng L, Ma J, Chi W et al (2006) LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18:955–969
Pérezpérez JM, Serralbo O, Vanstraelen M et al (2008) Specialization of CDC27 function in the Arabidopsis thaliana anaphase-promoting complex (APC/C). Plant J 53:78–89
Ponting CP (2000) Proteins of the endoplasmic-reticulum-associated degradation pathway: domain detection and function prediction. Biochem J 351(Pt 2):527–535
Prasad BD, Goel S, Krishna P (2010) In Silico identification of Carboxylate clamp type Tetratricopeptide repeat proteins in Arabidopsis and Rice as putative co-chaperones of Hsp90/Hsp70. PLoS One 5:e12761
Qbadou S, Becker T, Mirus O et al (2006) The molecular chaperone Hsp90 delivers precursor proteins to the chloroplast import receptor Toc64. EMBO J 25:1836–1847
Qin F, Kodaira K, Maruyama K et al (2011) SPINDLY, a negative regulator of Gibberellic acid Signaling, is involved in the plant Abiotic stress response. Plant Physiol 157:1900–1913
Ramon NM, Bartel B (2010) Interdependence of the Peroxisome-targeting receptors in Arabidopsis Thaliana: PEX7 facilitates PEX5 accumulation and import of PTS1 cargo into Peroxisomes. Mol Biol Cell 21:1263–1271
Rosado A, Schapire AL, Bressan RA et al (2006) The Arabidopsis tetratricopeptide repeat-containing protein TTL1 is required for osmotic stress responses and abscisic acid sensitivity. Plant Physiol 142:1113–1126
Rozas J, Sanchezdelbarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Schapire AL, Valpuesta V, Botella MA (2006) TPR proteins in plant hormone Signaling. Plant Signal Behav 1:229–230
Schweiger R, Muller NC, Schmitt MJ et al (2012) AtTPR7 is a chaperone docking protein of the sec translocon in Arabidopsis. J Cell Sci 125:5196–5207
Shao L (2012) Characterization of the TPR protein family and a putative photosynthetic protein from Synechocystis PCC 6803
She K, Kusano H, Koizumi K et al (2010) A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell 22:3280–3294
Sikorski RS, Boguski MS, Goebl M, Hieter P (1990) A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell 60:307–317
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Stockel J, Bennewitz S, Hein P, Oelmuller R (2006) The evolutionarily conserved tetratrico peptide repeat protein pale yellow Green7 is required for photosystem I accumulation in Arabidopsis and copurifies with the complex. Plant Physiol 141:870–878
Suzuki T, Nakajima S, Morikami A, Nakamura K (2005) An Arabidopsis protein with a novel calcium-binding repeat sequence interacts with TONSOKU/MGOUN3/BRUSHY1 involved in meristem maintenance. Plant Cell Physiol 46:1452–1461
Tang W, Kim T, Osesprieto JA et al (2008) BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321:557–560
Vonloeffelholz O, Kriechbaumer V, Ewan RA et al (2011) OEP61 is a chaperone receptor at the plastid outer envelope. Biochem J 438:143–153
Wang C, Tian Q, Hou Z et al (2007) The Arabidopsis thaliana AT PRP39-1 gene, encoding a tetratricopeptide repeat protein with similarity to the yeast pre-mRNA processing protein PRP39, affects flowering time. Plant Cell Rep 26:1357–1366
Wei K, Han P (2016) Pentatricopeptide repeat proteins in maize. Mol Breed 36:170
Wei K, Pan S (2014) Maize protein phosphatase gene family: identification and molecular characterization. BMC Genomics 15:773
Xie F, Zhang B (2010) Target-align: a tool for plant microRNA target identification. Bioinformatics 26:3002–3003
Yan J, Wang J, Li Q et al (2003) AtCHIP, a U-box-containing E3 ubiquitin ligase, plays a critical role in temperature stress tolerance in Arabidopsis. Plant Physiol 132:861–869
Yang SF, Hoffman NE (1984) Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol 35:155–189
Yang Z, Gu S, Wang X et al (2008) Molecular evolution of the CPP-like gene family in plants: insights from comparative genomics of Arabidopsis and Rice. J Mol Evol 67:266–277
Yang H, Liao L, Bo T et al (2014) Slr0151 in Synechocystis sp. PCC 6803 is required for efficient repair of photosystem II under high-light condition. J Integr Plant Biol 56:1136–1150
Young JC, Hoogenraad NJ, Hartl FU (2003) Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112:41–50
Zentella R, Hu J, Hsieh WP et al (2016) O-GlcNAcylation of master growth repressor DELLA by SECRET AGENT modulates multiple signaling pathways in Arabidopsis. Genes Dev 30:164–176
Zheng B, Chen X, Mccormick S (2011) The anaphase-promoting complex is a dual integrator that regulates both MicroRNA-mediated transcriptional regulation of Cyclin B1 and degradation of Cyclin B1 during Arabidopsis male gametophyte development. Plant Cell 23:1033–1046
Acknowledgments
We are grateful to the providers who submitted microarray and RNA-seq data to the public expression databases, which can be freely applied.
Funding
The project was supported by the Science and Technology Cooperation Project of Fujian Province, China (Grant No.2015I0006).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Table S1
The identified TPR proteins and their related information. 1 Isoelectric Point of TPRs. 2 Molecular Weight of TPRs. 3 Probability of export to mitochondria and chloroplast. 4 Predicted subcellular localization of TPRs. “C”, “M”, “S”, and “_” are represent as “chloroplast”, “mitochondrion”, “secretory pathway” and “any other location”, respectively. (PDF 461 kb)
Table S2
TPR protein information for polar (Populus trichocarpa). (PDF 105 kb)
Table S3
The Ka/Ks ratios and estimate of the absolute dates for the duplication events between the duplicated TPR gene pairs. (PDF 85 kb)
Table S4
The probe sets of AtTPR in this study. (PDF 33 kb)
Table S5
The normalized expression values (log2-based) of 161 Arabidopsis TPR genes in 63 tissues and developmental stages. (PDF 387 kb)
Table S6
The expression values of 125 OsTPR genes in 62 different tissues and development stages. (PDF 367 kb)
Table S7
List of 192 detected ZmTPR genes and their expression values in sixty distinct tissues in inbred line B73. (PDF 457 kb)
Table S8
Cis-acting elements were predicted from 1.5 kb upstream promoter region of the TPRs. (PDF 27 kb)
Table S9
OsTPR and ZmTPR expression patterns of successive stages of leaf development. (PDF 170 kb)
Table S10
The transcription levels of OsTPRs under drought stress in rice. (PDF 45 kb)
Table S11
The expression values of ZmTPRs in drought-stressed leaf and cob. MLC and MLD stand for well-watered control and drought stressed leaves, respectively, while MCC and MCD means well-watered control and drought stressed cobs, respectively. Numbers 1 and 2 indicate the two biological replicates. The extent of differential expression is measured in terms of fold change and (−) indicates failure to calculate or undetected values. Values highlighted in red and blue represent the expression levels of up-regulated and down-regulated genes, respectively. (PDF 99 kb)
Table S12
Primers used in this study. (PDF 11 kb)
Table S13
TPR genes and their regulatory miRNAs in Arabidopsis, rice and maize. (PDF 225 kb)
Fig. S1
Circos diagram of TPR gene pairs among Arabidopsis, rice and maize genomes. Outer two circles showed the distribution of each of the TPR genes and scaled chromosomes for each species in megabase (Mb) units, respectively. Histograms below each chromosome were the value of molecular weights (MWs) of TPR genes (blue <80, yellow ≥100); the two line plots were the isoelectric points (pIs) (green <7, yellow ≥7) and the repeat numbers of each TPRs (green <3, orange ≥16). (PDF 216 kb)
Fig. S2
An ML phylogenetic tree of TPR genes in Synechocystis PCC 6803, Arabidopsis, rice, maize and poplar. The TPR genes are grouped into eleven distinct groups (A-K). The gene names marked with the same colors belong to the same subfamilies, and the branch lines in different colors represent different species. Intron numbers of each TPR genes are shown on the right side. (PDF 481 kb)
Fig. S3
The map of exon/intron arrangement of TPR genes in Arabidopsis, rice and maize. (PDF 926 kb)
Fig. S4
Genomic distribution of AtTPR, OsTPR and ZmTPR genes on chromosomes. Different subfamilies of TPR genes were marked by different colors. (PDF 447 kb)
Fig. S5
Chromosomal segments containing TPR genes from Arabidopsis, rice and maize genomes. The Arabidopsis, rice and maize genomes are abbreviated as At, Os and Zm, respectively. Regions that putatively correspond to homologous genome blocks are connected by gray lines. (PDF 417 kb)
Fig. S6
The structure features of ZmTPR080. a Sequence alignment of eight motifs of ZmTPR080 (red, conserved residues; and blue, hydrophobic residues). Secondary structure of a typical TPR motif is shown above the alignment, with gray and purple bars representing helices. b Ramachandran plot and ERRAT result of ZmTPR080 homology structure. (PDF 553 kb)
Fig. S7
Hierarchial clustering display of TPR transcripts levels detected in distinct tissues in Arabidopsis, rice and maize. (PDF 855 kb)
Fig. S8
Expression profiling of TPR genes along rice and maize leaf developmental gradients. (PDF 467 kb)
Fig. S9
Pearson’s correlation of rice and maize gene expression based on 99 orthologous gene pairs. (PDF 461 kb) (PDF 389 kb)
Rights and permissions
About this article
Cite this article
Wei, K., Han, P. Comparative functional genomics of the TPR gene family in Arabidopsis, rice and maize. Mol Breeding 37, 152 (2017). https://doi.org/10.1007/s11032-017-0751-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-017-0751-4