Abstract
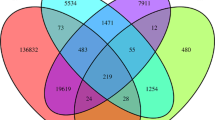
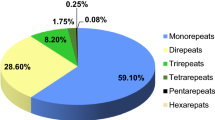
Mango (Mangifera indica) is the most important horticultural fruit crop in India, but few genetic markers have been identified in it. In order to develop genomic marker resources for mango, we sequenced genomic DNA using next-generation sequencing technology on the IlluminaHiSeq 2000 platform and examined sequence data for microsatellite markers. High-quality raw data were assembled and 198,612 contigs were obtained after optimization. From these data, 159,228 scaffolds were generated covering a genome size of 253.6 Mbp. From the scaffolds, 106,049 microsatellite repeats were identified. Finally, we were able to design primers for 84,118 microsatellites. Ninety simple sequence repeat (SSR) markers were tested, employing 64 mango cultivars and four Mangifera species, for determination of polymorphism and cross-species amplification. We identified 2103 alleles, and the allele number per locus ranged from 15 to 36. The majority of these markers amplified DNA in related species with a transferability of 94.4–98.8 %. The present study increases the sequence coverage of the mango genome and the number of mango-specific SSR markers. This is also the first report of the development of genomic SSR markers in mango using next-generation sequence technology. The genomic SSR markers identified in this study will be useful in diversity, identification, mapping and breeding studies.
Similar content being viewed by others
References
Arumuganathan K, Earle ED (1991) Estimation of nuclear DNA content of plants by flow cytometry. Plant Mol Biol Report 9:221–231
Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W (2011) Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27(4):578–579
Bohra A, Dubey A, Saxena RK, Penmetsa RV, Poornima KN (2011) Analysis of BAC-end sequences (BESs) and development of BES-SSR markers for genetic mapping and hybrid purity assessment development in pigeonpea (Cajanus spp.). BMC Plant Biol 11:56
Buschiazzo E, Gemmell NJ (2006) The rise, fall and renaissance of microsatellites in eukaryotic genomes. Bioessays 28(10):1040–1050
Cavagnaro PF, Senalik DA, Yang L, Simon PW, Harkins TT, Kodira CD, Huang S, Weng Y (2010) Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.). BMC Genom 11:569–586
Celik I, Gultekin V, Allmer J, Doganlar S, Frary A (2014) Development of genomic simple sequence repeat markers in opium poppy by next-generation sequencing. Mol Breed 34:323–334
Chiang YC, Tsai CM, Chen YKH, Lee SR, Chen CH, Lin YS, Tsai CC (2012) Development and characterization of 20 new polymorphic microsatellite markers from Mangifera indica (Anacardiaceae). Am J Bot 99:e117–e119
Daniel RZ, Ewan B (2008) VELVET: algorithm for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi:10.1101/gr.074492.107
Dutta S, Kumawat G, Singh BP, Gupta DK, Singh S, Dogra V, Gaikwad K, Sharma TR, Raje RS, Bandhopadhya TK, Datta S, Singh MN, Bashasab F, Kulwal P, Wanjari KB, Varshney RK, Cook DR, Singh NK (2011) Development of genic-SSR markers by deep transcriptome sequencing in pigeonpea [Cajanus cajan (L.) Millspaugh]. BMC Plant Biol 11:17
Duval MF, Bunel J, Sitbon C, Risterucci AM (2005) Development of microsatellite markers of mango. Mol Ecol Notes 5:824–826
Glenn TC, Schable TC (2005) Isolating microsatellite DNA loci. Methods Enzymol 395:202–222
Honsho C, Nishiyama K, Eiadthong W, Yonemori K (2005) Isolation and characterization of new microsatellites markers in mango. Mol Ecol Notes 5:152–154
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099–1106
Odeny DA, Jayashree B, Gebhardt Crouch J (2009) New microsatellite markers for pigeonpea (Cajanus cajan (L.) Millsp.). BMC Res Notes 2:35. doi:10.1186/1756-0500-2-35
Ratnaparkhe MB, Gupta VS, Murthy MRV, Ranjekar PK (1995) Genetic fingerprinting of pigeonpea [Cajanus cajan (L.) Millsp.] and wild relatives using RAPD markers. Theor Appl Genet 91:893–898
Ravishankar KV, Anand L, Dinesh MR (2000) Assessment of genetic relatedness among a few Indian mango cultivars using RAPD markers. J Hortic Sci Biotechnol 75:198–201
Ravishankar KV, Mani BH, Anand L, Dinesh MR (2011) Development of new microsatellite markers from mango (Mangifera indica) and cross-species amplification. Am J Bot 98(4):e96–e99
Schnell RJ, Olano CT, Quintanilla WE, Meerow AW (2005) Isolation and characterization of 15 microsatellite loci from mango (Mangifera indica L.) and cross-species amplification in closely related taxa. Mol Ecol Notes 5:625–627
Schuelke M (2000) An economic method for the fluorescent labelling of PCR fragments. Nat Biotechnol 18:233–234
Selale H, Celik I, Gultekin V, Allmer J, Doganlar S, Frary A (2013) Development of EST-SSR markers for diversity and breeding studies in opium poppy (Papaver somniferum L.). Plant Breed 132:344–351. doi:10.1111/pbr.12059
Sonah H, Deshmukh RK, Sharma A, Sing VP, Gupta DK, Gacche RN, Rana JC, Singh NK, Sharma TR (2011) Genome-wide distribution and organization of microsatellites in plants: an insight into marker development in Brachypodium. PLoS ONE 6(6):e21298:1–e21298:9
Steve R, Skaletsky HJ (1996) Primer 3. http://wwwgenome.wi.mit.edu/genome_software/other/Primer3.html
Ukoskit K (2007) Development of microsatellite markers in mango (Mangifera indica L.) using 5′ anchored PCR. Thammasat Int J Sci Technol 12:3
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellites markers in plants: features and application. Trends Biotechnol 23:48–55
Viruel MA, Escribano P, Barbieri M, Ferri M, Hormaza JI (2005) Fingerprinting, embryo type and geographic differentiation in mango (Mangifera indica L., Anacardiaceae) with microsatellites. Mol Breed 15:383–393. doi:10.1007/s11032-004-7982-x
Yang SY, Pang W, Ash G, Harper J, Carling J, Wenzl P, Huttner E, Zong XX, Kilian A (2006) Low level of genetic diversity in cultivated pigeon pea compared to its wild relatives is revealed by diversity arrays technology. Theor Appl Genet 113:585–595
Zalapa JE, Cuevas H, Zhu H, Steffan S, Senalik D, Zeldin E, McCown B, Harbut R, Simon P (2012) Using next-generation sequencing approaches to isolate simple sequence repeat (SSR) loci in the plant sciences. Am J Bot 99:193–208
Acknowledgments
K.V.R. and M.R.D. acknowledge financial support from Department of Biotechnology, New Delhi funded projects “Morphological and Molecular Characterization of Wild and Indigenous Mango Varieties of Indo-Burma Region (North-Eastern Region) and National Database on Mango”.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ravishankar, K.V., Dinesh, M.R., Nischita, P. et al. Development and characterization of microsatellite markers in mango (Mangifera indica) using next-generation sequencing technology and their transferability across species. Mol Breeding 35, 93 (2015). https://doi.org/10.1007/s11032-015-0289-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-015-0289-2