Abstract
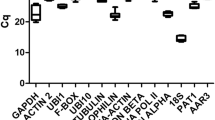
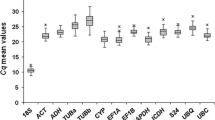
Normalisation to a reference gene is the most common method of internally controlling for error in quantitative PCR (qPCR) experiments. Studies based on qPCR in chickpea have been carried out using potential reference genes exclusively. Inappropriate normalisation may result in the acquisition of biologically irrelevant data. We have tested the expression of 12 candidate internal control genes in 36 samples representing different organs/developmental stages, genotypes and stress conditions. The most stably expressed genes were PUBQ, GAPDH, UBQ and bHLH, whereas 18S rRNA and EF-1a showed considerable regulation. The most suitable combination of reference genes for the particular experimental sets tested is provided. To illustrate the use of chickpea reference genes, we checked the expression of a putative defence gene in two different genotypes infected with Ascochyta rabiei (Pass.) Lab. The set of reference genes presented here will enable the more accurate and reliable normalisation of qPCR results for gene expression studies in this important legume crop. Our findings can be used as a starting point for reference gene selection in experimental conditions different from those tested here.



Similar content being viewed by others
References
Ashraf N, Ghai D, Barman P, Basu S, Gangisetty N, Mandal MK, Chakraborty N, Datta A, Chakraborty S (2009) Comparative analyses of genotype dependent expressed sequence tags and stress-responsive transcriptome of chickpea wilt illustrate predicted and unexpected genes and novel regulators of plant immunity. BMC Genomics 10:415
Bustin SA (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25:169–193
Bustin SA (2002) Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol 29:23–39
Bustin SA (2010) Why the need for qPCR publication guidelines?—The case for MIQUE. Methods 50:217–226
Bustin SA, Nolan T (2004) Pitfalls of quantitative real-time reverse transcription polymerase chain reaction. J Biomol Tech 15:155–166
Bustin SA, Benes V, Nolan T, Pfaffl MW (2005) Quantitative real-time RT-PCR—a perspective. J Mol Endocrinol 34:597–601
Bustin SA, Benes V, Garson JA, Hellemans J, Huguett J, Kubista M, Mueller R et al (2009) The MIQE Guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622
Cho S, Muehlbauer FJ (2004) Genetic effect of differentially regulated fungal response genes on resistance to necrotrophic fungal pathogens in chickpea (Cicer arietinum L.). Physiol Mol Plant Pathol 64:57–66
Cruz F, Kalaoun S, Nobile P, Colombo C, Almeida J, Barros L, Romano E et al (2009) Evaluation of coffee reference genes for relative expression studies by quantitative real-time RT-PCR. Mol Breed 23:607–616
Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38:366–379
Czechowski T, Stitt M, Altman T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17
Derveaux S, Vandesompele J, Hellemans J (2010) How to do successful gene expression analysis using real-time PCR. Methods 50:227–230
Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, Zumla A (2004) Validation of housekeeping genes for normalizing RNA expression in real-time PCR. BioTechniques 37:112–119
Dheda K, Huggett JF, Chang JS, Kim LU, Bustin SA, Johnson MA, Rook GAW, Zumla A (2005) The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal Biochem 344:141–143
Die JV, Dita MA, Krajinski F, González-Verdejo CI, Rubiales D, Moreno MT, Román B (2007) Identification by suppression subtractive hybridization and expression analysis of Medicago truncatula putative defence genes in response to Orobanche crenata parasitization. Physiol Mol Plant Pathol 70:49–59
Dixon RA, Sumner LW (2003) Legume natural products: Understanding and manipulating complex pathways for human and animal health. Plant Physiol 131:878–885
Dixon RA, Acnine L, Kota P, Liu CJ, Reddy MSS, Wang LJ (2002) The phenilpropanoid pathway and plant defence—a genomics perspective. Mol Plant Pathol 3:371–390
Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Pérez JA (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8:131
Filby AL, Tyler CR (2007) Appropriate ‘housekeeping’ genes for use in expression profiling the effects of environmental estrogens in fish. BMC Mol Biol 8:10
Fleige S, Pfaffl MW (2006) RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med 27:126–139
Fleige S, Walf V, Huch S, Prgomet C, Sehm J, Pfaffl MW (2006) Comparison of relative mRNA quantification models and the impact of RNA integrity in quantitative real-time RT- PCR. Biotechnol Lett 28:1601–1613
Food and Agriculture Organization (2008) FAO Statistical Databases—Agricultural Production (updated: 19 December 2009). URL: http://apps.fao.org/page/collections. Accessed on 20 June 2010
Frost P, Nilsen F (2003) Validation of reference genes for transcription profiling in the salmon louse, Lepeophtheirus salmonis, by quantitative real-time PCR. Vet Parasitol 118:169–174
Gachon C, Mingam A, Charrier B (2004) Real-time PCR: what relevance to plant studies? J Exp Bot 55:1445–1454
Garg R, Sahoo A, Tyagi AK, Jain M (2010) Validation of internal control genes for quantitative gene expression studies in chickpea (Cicer arietinum L.). Biochem Biophys Res Commun. doi:10.1016/j.bbrc.2010.04.079
Guénin S, Mauriat M, Pelloux J, Van Wuytswinkel O, Bellini C, Gutierrez L (2009) Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. J Exp Bot 60:487–493
Gutierrez L, Mauriat M, Pelloux J, Bellini C, Van Wuytswinkel O (2008a) Towards a systematic validation of references in real-time RT-PCR. Plant Cell 20:1734–1735
Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, Van Wuytswinkel O (2008b) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotech J 6:609–618
Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8:R19
Hoebeeck J, van der Luitj R, Poppe B, De Smet E, Yigit N, Claes K et al (2005) Rapid detection of VHL exon deletions using real-time quantitative PCR. Lab Invest 85:24–33
Hoebeeck J, Speleman F, Vandesompele J (2007) Real-time quantitative PCR as an alternative to southern blot or fluorescence in situ hybridization for detection of gene copy number number changes. In: Hilario E, Mackay J (eds) Methods in Molecular Biology, vol 353. Humana Press Inc, Totowa, NJ, pp 205–225
Hu R, Fan C, Li H, Zhang Q, Fu Y-F (2009) Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol Biol 10:93
Hugget J, Dheda K, Bustin S, Zumla A (2005) Real-time RT-PCR normalisation; strategies and considerations. Genes Immun 6:279–284
Iruela M, Castro P, Rubio J, Cubero JI, Jacinto C, Millán T, Gil J (2007) Validation of a QTL for resistance to ascochyta blight linked to resistance to fusarium wilt race 5 in chickpea (Cicer arietinum L.). Eur J Plant Pathol 119:29–37
Kakar K, Wandrey M, Czechowski T, Gaertner T, Scheible W-R, Stitt M, Torres-Jerez I et al (2008) A community resource for high-throughput quantitative RT-PCR analysis of transcription factor gene expression in Medicago truncatula. Plant Methods 4:18
Kaur G, Kumar S, Nayyar H, Upadhyaya HD (2008) Cold stress injury during the pod-filling phase in chickpea (Cicer arietinum L.): effects on quantitative and qualitative components of seeds. J Agron Crop Sci 194:457–464
Lefever S, Vandesompele J, Speleman F, Pattyn F (2009) RTPrimerDB: the portal for real-time PCR primers and probes. Nucleic Acids Res 37:D942–D945
Liu CJ, Huhman D, Summer LW, Dixon RA (2003) Regiospecific hydroxylation of isoflavones by cytochrome p450 81E enzymes from Medicago truncatula. Plant J 36:471–484
Mantri NL, Ford R, Coram TE, Pang ECK (2007) Transcriptional profiling of chickpea genes differentially regulated in response to high-salinity, cold and drought. BMC Genomics 8:303
Millan T, Clarke H, Siddique K, Buhariwalla H, Hutokshi G (2006) Chickpea molecular breeding: new tools and concepts. Euphytica 147:81–103
Munns R, Husain S, Rivelli A, James R, Condon A, Lindsay M, Lagudah E, Schachtman D, Hare R (2002) Avenues for increasing salt tolerance of crops, and the role of physiologically based selection traits. Plant Soil 247:93–105
Navas-Cortés JA, Pérez-Artés E, Jiménez-Diaz RM, Llobell A, Bainbridge BW, Heale JB (1998) Mating type, pathotype, and RAPDs analysis in Didymella rabiei, the agent of ascochyta blight of chickpea. Phytoparasitica 26:199–212
Nayyar H, Kaur G, Kumar S, Upadhyaya HD (2007) Low temperature effects during seed filling on chickpea genotypes (Cicer arietinum L.): probing mechanisms affecting seed reserves and yield. J Agron Crop Sci 193:336–344
Nolan T, Hands RE, Bustin SA (2006) Quantification of mRNA using real-time RT-PCR. Nat Protoc 1:1559–1582
Overkamp S, Hein F, Barz W (2000) Cloning and characterization of eight cytochrome P450 cDNAs from chickpea (Cicer arietinum L.) cell suspension cultures. Plant Sci 155:101–108
Perez-Novo CA, Claeys C, Speleman F, Cauwenberge PV, Bachert C, Vandesompele J (2005) Impact of RNA quality on reference gene expression stability. BioTechniques 39:52–56
Pfaffl MW (2006) Relative quantification. In: T Dorak (ed) Real-time PCR. pp 63–82
Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A (2004) Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun 23:856–862
Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 13:62–66
Remans T, Smeets K, Opdenakker K, Mathijsen D, Vangronsveld J, Cuypers A (2008) Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 227:1343–1349
Robinson TL, Sutherland IA, Sutherland J (2007) Validation of candidate bovine reference genes for use with real-time PCR. Vet Immunol Immunopathol 115:160–165
Shimada N, Akashi T, Aoki T, Ayabe S (2000) Induction of isoflavonoid pathway in the model legume Lotus japonicus: molecular characterization of enzymes involved in phytoalexin biosynthesis. Plant Sci 160:37–47
Singh KB, Omar M, Saxena MC, Johansen C (1997) Screening for drought resistance in spring chickpea in the mediterranean region. J Agron Crop Sci 178:227–235
Srinivasan A, Johansen C, Saxena NP (1998) Cold tolerance during early reproductive growth of chickpea (Cicer arietinum L.): characterization of stress and genotypic variation in pod set. Field Crops Res 57(2):181–193
Udvardi MK, Czechowski T, Scheible W-R (2008) Eleven golden rules of quantitative RT-PCR. Plant Cell 20:1736–1737
Vadez V, Krishnamurthy L, Serraj R, Gaur PM, Upadhyaya HD, Hoisington DA, Varshney RK, Turner NC, Siddique KHM (2007) Large variation in salinity tolerance in chickpea is explained by differences in sensitivity at the reproductive stage. Field Crops Res 104:123–129
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002a) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034
Vandesompele J, De Paepe A, Speleman F (2002b) Elimination of primer–dimer artifacts and genomic coamplification using a two-step SYBR Green I real-time RT-PCR. Anal Biochem 303:95–98
VanGuilder HD, Vrana KE, Freeman WM (2008) Twenty-five years of quantitative PCR for gene expression analysis. BioTechniques 44:619–626
Varshney RK, Close TJ, Singh NK, Hoisington DA, Cook DR (2009) Orphan legume crops enter the genomics era! Curr Opin Plant Biol 12:202–210
Wong ML, Medrano JF (2005) Real-time PCR for mRNA quantitation. BioTechniques 39:75–85
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31(13):3406–3415
Acknowledgments
Financial support was provided by the Spanish project RTA2007-00009. José V. Die is a researcher funded by the ‘Juan de la Cierva’ programme of the Spanish Ministerio de Ciencia e Innovación. We thank E. Madrid for providing us with primer cyp81E3 sequence.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Castro, P., Román, B., Rubio, J. et al. Selection of reference genes for expression studies in Cicer arietinum L.: analysis of cyp81E3 gene expression against Ascochyta rabiei . Mol Breeding 29, 261–274 (2012). https://doi.org/10.1007/s11032-010-9544-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-010-9544-8