Abstract
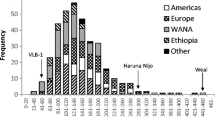
Five single nucleotide polymorphism (SNP) sites corresponding to substitutions in the protein sequence of the β-amylase gene Bmy1 at amino acid (AA) positions 115, 165, 233, 347 and 430 were genotyped in 493 cultivated barley accessions by Pyrosequencing and a CAPS assay. A total of 6 different haplotypes for the Bmy1 gene were discovered of which 4 haplotypes were identified as previously described alleles Bmy1-Sd1, Bmy1-Sd2L, Bmy1-Sd2H and Bmy1-Sd3, while 2 haplotypes were new. A broad spectrum of haplotypes was found in spring barleys, while the winter barleys were dominated by the newly described haplotype Bmy1-Sd4. Individual haplotype frequencies varied between the geographic regions.
Three pairs of SNP loci within the gene showed highly significant (P<0.0001) elevated values of linkage disequilibrium (LD) with r 2 > 0.6. In the European and Asian subpopulations different loci were in linkage disequilibrium due to the differences in haplotype frequency distributions. By applying LD data to select haplotype tagging SNPs, three SNP sites corresponding to AA positions 115, 233 and 347 were identified that allowed to discriminate 4 haplotypes and to capture 91.6% of the available diversity by distinguishing 452 out of 493 accessions.
In a subset of 2-rowed German spring barley varieties 4 SNPs and 2 haplotypes had a significant association with the malting quality parameter final attenuation limit which is related to the total amylolytic enzymatic activity.
Similar content being viewed by others
References
Allison MJ, Swanston JS (1974) Relationships between b-amylase polymorphisms in developing, mature and germinating grains of barley. J Inst Brew 80:285–291
Beschreibende Sortenliste. Getreide, Mais, Ölfrüchte, Leguminosen, Hackfrüchte. Bundessortenamt. 1994–2003. Landbuch Verlagsgesellschaft mbH. Hannover
Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA (2004) Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet 74:106–120
Clark SE, Hayes PM, Henson CA (2003) Effects of single nucleotide polymorphisms in β-amylase1 alleles from barley on functional properties of the enzymes. Plant Physiol Biochem 41(9):798–804
Clancey JA, Han F, Ullrich SE (2003) Comparative mapping of β-amylase activity QTLs among three barley crosses. Crop Sci 43:1043–1052
Coventry SJ, Collins HM, Barr AR, Jefferies SP, Chalmers KJ, Logue SJ, Langridge P (2003) Use of putative QTLs and structural genes in marker assisted selection for diastatic power in malting barley (Hordeum vulgare L). Aust J Agr Res 54:1241–1250
Dubcovsky J, Chen C, Yan L (2005) Moleculat characterization of the allelic variation at the VRN-H2 vernalization locus in barley. Mol Breeding 15:395–407
Eglinton JK, Langridge P, Evans DE (1998) Thermostability variation in alleles of barley β-amylase. J Cereal Sci 28:301–309
Erkkilä MJ, Ahokas H (2001) Special barley β-amylase allele in a Finnish landrace line HA52 with high grain enzyme activity. Hereditas 134:91–95
Erkkilä MJ, Leah R, Ahokas H, Cameron-Mills V (1998) Allele-dependent barley grain β-amylase activity. Plant Physiol 117:679–685
Franckowiak J (1997) Revised linkage maps for morphological markers in barley, Hordeum vulgare. Barley Genetics Newsletter 26:9–21
Kaneko T, Kihara M, Ito K (2000) Genetic analysis of β -amylase thermostability to develop a DNA marker for malt fermentability improvement in barley, Hordeum vulgare. Plant Breeding 119:197–201
Kihara M, Kaneko T, Ito K (1998) Genetic variation of β-amylase thermostability among varieties of barley, Hordeum vulgare L., and relation to malting quality. Plant Breeding 117:425–428
Kreis M, Williamson M, Buxton B, Pywell J, Hejgaard J, Svendsen I (1987) Primary structure and differential expression of β-amylase in normal and mutant barley. Eur J Biochem 169:517–525
Kreis M, Williamson M, Shewry PR, Sharp P, Gale M (1988) Identification of a 2nd locus encoding β-amylase on chromosome-2 of barley. Genetical Res 51:13–16
Laurie DA, Pratchett N, Bezant JH, Snape JW (1995) RFLP mapping of 5 major genes and 8 quantitative trait loci controlling flowering time in a winter x spring barley (Hordeum vulgare L.) cross. Genome 38(3):575–585
Li C, Langridge P, Zhang X, Eckstein PE, Rossnagel BG, Lance RC, Lefol EB, Lu M, Harvey BL, Scoles GJ (2000) Hordeum vulgare cultivar RFLP-Harrington Bmy1 gene for Sd1 beta-amylase, complete cds. Direct submission to GenBank AB048949
Lin J-Z, Morell PL, Clegg MT (2002) The influence of linkage and inbreeding on patterns of nucleotide sequence diversity at duplicate alcohol dehydrogenase loci in wild barley (Hordeum vulgare ssp. spontaneum). Genetics 162:2007–2015
Ma YF, Evans DE, Logue SJ, Langridge P (2001) Mutations of barley β-amylase that improve substrate-binding affinity and thermostability. Mol Genet Genomics 266:345–52
Malysheva L, Ganal MW, Röder MS (2004) Evaluation of cultivated barley (Hordeum vulgare L.) germplasm for the presence of thermostable alleles of β-amylase. Plant Breeding 123:128–131
Malysheva-Otto L, Ganal MW, Röder MS (2006) Analysis of molecular diversity, population structure and linkage disequilibrium in worldwide survey of cultivated barley germplasm (Hordeum vulgare L.). BMC. Genetics 7:6
Massa AN, Morris CF, Gill BS (2004) Sequence diversity of Puroindoline-a, Puroindoline-b, and the grain softness protein genes in Aegilops tauschii Coss. Crop Sci 44:1808–1816
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76(10):5269–5273
Palaisa K, Morgante M, Tingey S, Rafalski A (2004) Long-range pattern of diversity and linkage disequilibrium surrounding the maize gene Y1 gene are indicative of an asymmetric selective sweep. Proc Natl Acad Sci USA 101(26):9885–90
Paris M, Jones MGK, Eglinton JK (2002) Genotyping single nucleotide polymorphisms for selection of barley β-amylase alleles. Plant Mol Biol Rep 20:149–159
Plaschke J, Ganal MW, Röder MS (1995) Detection of genetic diversity in closely related bread wheat using microsatellite markers. Theor Appl Genet 91:1001–1007
Polakova K, Laurie D, Vaculova K, Ovesna J (2003) Characterization of β-amylase alleles in 79 barley varieties with Pyrosequencing. Plant Mol Biol Rep 21:439–47
Potokina E, Prasad M, Malysheva L, Röder MS, Graner A (2006) Expression genetics and haplotype analysis reveal cis regulation of serine carboxypeptidase I (Cxp1), a candidate gene for malting quality in barley (Hordeum vulgare L.). Func Integr Genomics 6: 25–35
Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM, Buckler ESIY (2001) Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Natl Acad Sci USA 98(20):11479–84
Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier M-H, Leroy P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Rohlf FJ (1998) NTSYS-pc: numerical taxonomy and multivariate analysis system. Applied Biostatistics Inc, New York
Sjakste TG, Röder M (2004) Distribution and inheritance of β-amylase alleles in north European barley varieties. Hereditas 141:39–45
Szalma SJ, Buckler ES IV, Snook ME, McMullen MD (2005) Association analysis of candidate genes for maysin and chlorogenic acid accumulation in maize silks. Theor Appl Genet 110:1324–1333
Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielse D, Buckler IV ES (2001) Dwarf8 polymorphisms associate with variation in flowering time. Nature Genetics 28:286–289
von Wettstein-Knowles P (1993) Barley (Hordeum vulgare) 2N = 14. In: Genetic maps: locus maps of complex genomes. Cold Spring Harbor Laboratory Press 1993 Ed. O’Brien SJ
Yoshigi N, Okada Y, Sahara H, Koshino S (1994) PCR cloning and sequencing of the beta-amylase cDNA from barley. J Biochem (Tokyo) 115:47–51
Yoshigi N, Okada Y, Sahara H, Tamaki T (1995) A structural gene encoding beta-amylase of barley. Biosci Biotechnol Biochem 10:1991–1993
Zhang W, Kaneko T, Ishii M, Takeda K (2004) Differentiation of β-amylase phenotypes in cultivated barley. Crop Sci 44:1608–1614
Acknowledgement
We thank Ellen Weiß for excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Malysheva-Otto, L., Röder, M. Haplotype diversity in the endosperm specific β-amylase gene Bmy1 of cultivated barley (Hordeum vulgare L.). Mol Breeding 18, 143–156 (2006). https://doi.org/10.1007/s11032-006-9023-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-006-9023-4